Keywords
Computer Science and Digital Science
- A3.1.1. Modeling, representation
- A3.4.4. Optimization and learning
- A3.4.5. Bayesian methods
- A6.1.1. Continuous Modeling (PDE, ODE)
- A6.1.2. Stochastic Modeling
- A6.1.4. Multiscale modeling
- A6.3.1. Inverse problems
- A6.3.3. Data processing
- A6.4.1. Deterministic control
- A6.4.3. Observability and Controlability
Other Research Topics and Application Domains
- B1.1.2. Molecular and cellular biology
- B1.1.7. Bioinformatics
- B1.1.8. Mathematical biology
- B1.1.10. Systems and synthetic biology
- B2.4.2. Drug resistance
- B5.10. Biotechnology
- B9.8. Reproducibility
1 Team members, visitors, external collaborators
Research Scientists
- Grégory Batt [Team leader, Inria, Senior Researcher, HDR]
- Jakob Ruess [Inria, Researcher]
Post-Doctoral Fellows
- Virgile Andreani [Institut Pasteur, from Dec 2020]
- Olivier Borkowski [Inria]
- Zachary Fox [Inria, until Oct 2020]
- Davin Lunz [Inria, joint affiliataion with Commands Team]
PhD Students
- Chetan Aditya [Inria]
- Virgile Andreani [Inria and Institut Pasteur, until Nov 2020]
- Arthur Carcano [Université de Paris]
- Viktoriia Gross [Institut Pasteur, from Oct 2020]
- Sebastian Ramon Sosa Carrillo [Inria]
Technical Staff
- François Bertaux [Institut Pasteur, Engineer, Permanent Staff]
- Steven Fletcher [Institut Pasteur, Engineer, until Aug 2020]
- Achille Fraisse [Institut Pasteur, Engineer, from Sep 2020]
Visiting Scientist
- Lorenzo Pasotti [Università degli Studi di Pavia, Jan 2020 and Jul - Aug 2020, Assistant Professor]
2 Overall objectives
The main objective of our research is to understand, control, and optimize cellular processes in single cells and at the population level. We combine experimental and theoretical work within a single team.
Our focus is on developing methods and models that take stochasticity of intracellular processes and heterogeneity of cell populations into account. To this end, we use both mixed-effects models as well as continuous-time Markov chains and their diffusion approximations. We develop methods for efficiently calculating with such models and use them to design optimally informative experiments and to reverse engineer unknown cellular processes from experimental data. Furthermore, we deploy models in order to optimally construct and optimally control synthetic gene circuits.
We have recently started to set up our own biology laboratory at Institut Pasteur. We develop novel experimental platforms that are designed to be fully automated, controllable by our own software, and capabale of updating the experimental plan in response to incoming measurements. Optogenetic actuation of intracellular processes, coupled to real time fluorescence measurements by microscopy or flow cytometry, then allows us to connect cellular processes with models and algorithms in real time.
The spritit of our work is that experimental platforms and circuits should be constructed with our theoretical work in mind, while our mathematical methods should be usable to adress concrete experimental questions in the lab.
3 Research program
3.1 Analysis and identification of stochastic (biochemical) reaction networks
The advancement of single-cell technologies in the last decades revealed that stochasticity is an inherent feature of cellular processes. Stochastic models, governed by the chemical master equation (CME), are widely used in applications to shed light on the functioning of biochemical reaction networks inside single cells. However, in most cases, the analysis of such models is based exclusively on Gillespie’s stochastic simulation algorithm (SSA). SSA allows one to easily forward simulate the model but cannot be used very well for many important model analysis tasks. To overcome this problem, we develop various alternative approaches for calculating with stochastic models. In particular, we derive ordinary differential equations for the time evolutions of statistical moments of species abundances from the CME. We use moment equations and moment closure to develop methods for various model analysis tasks for which stochastic simulation is computationally too expensive to be practically useful, ranging from parameter inference to model predictive control. Furthermore, we recently started to make use of approximations of the CME with a Fokker-Planck equation to be able to also calculate with models for which low order statistical moments are not sufficient statistics of the full data and do not contain enough information.
3.2 Population dynamics emerging from randomness in single cells
Dynamics of cell populations growing in isolation or as part of some ecological system are often shaped by biochemical processes inside cells, for instance when these processes convey resistance to stressors or trigger cell fate decisions in reponse to environmental conditions. Understanding how stochastic reaction events inside single cells affect emerging population dynamics, and how selection effects at the population level feed back to shape single cell characteristics of cells in the population, is one of the key questions in biology. We develop multi-scale modeling approaches that allow us to derive emerging population dynamics from mechanistic descriptions of stochastic reaction networks inside single cells. In the past, we have used such approaches to study how stochasticity in restriction-modification systems, acting as simple bacterial innate immune systems, propagates to the ecology of bacteria and bacterial viruses and shapes the dynamics of bacterial populations. More generally, we develop and use these approaches in connection with experimental work in our lab for understanding and controlling the dynamics of populations in cases where controllable system inputs inherently operate at the level of single cells (e.g. optogenetics) but the output of interest is at the level of populations (e.g. bioproduction).
3.3 Optimal experimental design
One of the major problems in reverse engineering biochemical processes inside cells is that cellular processes are high-dimensional and complex with many unknown parameters while the available data is low dimensional and corrupted by measurement errors. Such problems can be alleviated by ensuring that the experimental plan is designed to yield data that provides as much information as possible about the unknown model parameters. We develop mathematical approaches and computational tools that can be used to calculate the expected amount of information that can be gained from a given experiment given a specification of either a stochastic model of the system (described above) or a deterministic model based on ordinary differential equations. These information calculation approaches are then coupled to optimization tools and used to plan maximally informative experiments in our applications.
3.4 Cybergenetics – real time control of biological processes
Cells have evolved uncountable numbers of feedback circuits to regulate their functionalities in the presence of changing environmental conditions. But can such feedback control also be externalized and placed under control of scientists? Early work on this topic suggested that optogenetic systems, allowing for external regulation of gene expression, have the potential to serve as an interface between cells and experimental platform that gives a computer the power to stir the functioning of cells via the application of light. We develop all the tools required to realize automated computer control of intracellular processes. On the experimental side, we develop yeast strains that are equipped with optogenetic promoters to drive various functionalities. On the mathematical side, we develop models and software to equip our experimental platforms with the appropriate programs to realize sucessful feedback control, both at the level of single cells (microscopy) and at the level of populations (bioreactors and plate reader).
3.5 Platforms for automated reactive experiments
The core scientific activity of the team is to connect mathematical methods with biological applications in our lab. The interface between the two sides, that is the experimental platforms, is therefore of crucial importance for the success of our activities. However, platforms that can be purchased by vendors are typically delivered without the capacity to adapt the experimental plan in response to incoming measurements, a functionality that is crucially needed for deploying our computational methods (e.g. feedback control). Therefore, we develop novel experimental platforms and/or extend existing platforms with additional software and hardware that allows us to perform automated reactive experiments. Concretely, we develop a microscopy platform and control software for yeast that uses a digital micromirror device to expose single cells to targeted light signals that can be adjusted in real time in response to measurements taken from the cell. Furthermore, we develop a platform of 16 parallel small scale automated bioreactors, each equipped with controllable LEDs to allow for optogenetic gene expression and long-term reactive experiments in tightly controlled conditions. Automation of the platform is achieved via a low-cost open-source pipetting robot that samples all reactors to a benchtop cytometer in which single cell gene expression is measured in all sampled cells of all reactors. Finally, we develop software to take full control of a commercial plate reader with liquid injection capabilities (Tecan Spark). This platform allows us to use a Raspberry Pi to pilot 96 parallel reactive experiments where optical density is used as a readout of bacterial growth.
4 Application domains
4.1 Preamble
Since most of our research is at the interface of mathematics and biology, there often is no clear split between mathematical reaseach objectives and applications. For instance, feedback control of gene expression is simultaneously a mathematical and an applied problem.
4.2 Understanding resistance and tolerance to antibiotic treatments
The non-susceptibility of pathogenic bacteria to antibiotic treatments is a major health problem. Bacteria might escape treatments in two ways: being resistant or being tolerant. Whereas resistant bacteria can multiply in presence of antibiotics, tolerant bacteria can merely survive. Yet, tolerance is increasingly recognized as a major player in treatment failure. In particular, an increasing fraction of commensal and pathogenic E coli bacteria express extended-spectrum β-lactamases and/or carbapenemases. When individual bacteria die as a consequence of antibiotic treatments, these enzymes are released and hydrolyze the antibiotic molecules in the environment, conveying a transient protection to the remaining bacteria that lasts until the enzymes are degraded themselves. Understanding how this collective antibiotic tolerance (CAT) shapes population dynamics is difficult yet important for optimally killing bacterial populations: when a second antibiotic dose is applied directly after a first dose it will not be effective since the antibiotics will be degraded by the enzymes released from bacteria killed after the first dose; when the second dose is applied too late the surviving bacterial population will have regrown to a large size. Our plate reader platform allows us to apply complex temporal patterns of antibiotic treatments to bacteria over nearly two days. Parallezing such treatments in the 96 well plates allows us to generate rich data sets and to calibrate population dynamics models that can be used to optimize temporal treatment plans. One of the applied objectives of our team is to use these capacities to study a collection of fully-sequenced clinical isolates treated with a broad range of clinically important antibiotics and grown in various media. Ideally, this will lead to an approach that can be used to assay tolerance to antibiotics in hospitals instead of, or in addition to, standard antibiotic susceptibility tests, detecting resistance.
4.3 Optimization of protein production in yeast
Many proteins are of technological or therapeutical importance. The yeast S. cerevisiae is an interesting organism for protein bioproduction since it combines a relatively fast growth rate with good capacities to perform post-translational modifications needed for protein maturation and full functionality. However, imposing a strong demand on protein production to the cell places a significant burden on its physiology, either at the protein production level or at the maturation and secretion levels. Using systems and synthetic biology approaches, we aim at better understanding the origins of the production bottlenecks and then using modeling and control approaches, we aim at finding optimal control solutions for bioproduction. Three different strategies are envisioned. In the first approach, bioproduction stress sensors are used to observe in real time the physiological state of the cell, and the demand is externally tuned based on the stress level of the cell population. In the second approach, the stress sensor is used to tune the response capacities of the cell to the external demand, thus creating an internal feedback loop. In the third approach, we control the fraction of the producing cells by engineering an artificial differentiation system that implements the partial differentiation of grower cells into producer cells. The optimization problem is then to find the optimum ratio based on the external environment of the cells.
5 Social and environmental responsibility
5.1 Footprint of research activities
A significant part of our daily research activities involves molecular biology work and consumes plasticware and various chemicals. We also work on lab automation and develop experimental platform to paralellize experiments. However, we work with small biorectors (15 to 50mL), so volumes of cell cultures remain very modest.
We also occasionally use a computer cluster, notably for optimization, but the jobs remain relatively modest on a yearly basis.
5.2 Impact of research results
Regarding biological developments, we have two main research directions.
The first one deals with the optimization of bioproduction. Bioproduction is a domain of strategic importance. The field is higly technological and rapidly growing at the global scale. The market for biopharmaceuticals alone, that notably include vaccines and monoclonal antibodies, is estimated to $400B to $500B. France imports of its vaccines and of its monoclonal antibodies and lacks sovereignty. Therefore this field has a strong social, medical and economical importance.
The second research direction deals with antibiotic stewardship. The spread of antimicrobial resistance is a both a health and an ecological problem of global impact. Antibiotic stewardship aims at using these drugs in more appropriate ways. To do so, one has to better understand and quantify bacterial response to antibiotic treatments.
Therefore our two main research directions are both tightly connected with important health and social issues.
6 New software and platforms
6.1 Reactive microscopy platforms with MicroMator
Participants: Steven Fletcher (main developper).
Software for microscopy automation are essential to support reproducible high-throughput microscopy experiments. Samples can now be routinely imaged using complex spatial and temporal patterns. Yet, in the overwhelming majority of the cases, executions of experiments are still cast in stone at the beginning. Software empowering microscopy with real-time adaptation capabilities is needed to exploit the full potential of automated microscopes.
To address this need, we developed MicroMator, a modular open-source Python software to support reactive microscopy. MicroMator is built upon and extends MicroManager, the default open-source cross-platform software for driving microscopes. The main difference with respect to the standard paradigm is that experiments are now defined by user-defined events, having trigger and action components. MicroMator is interfaced with complex hardware to form reactive microscopy platforms.
Several applications are also developped to demonstrate that MicroMator conveniently allows to employ complex experimental strategies that are currently at the forefront of microscopy automation research.
6.2 Automated and reactive bioreactor platforms with ReacSight
Participants: François Bertaux (main developper).
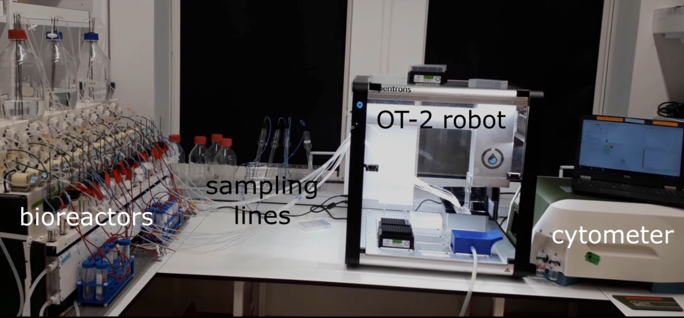
Small-scale, low-cost bioreactors are emerging as powerful tools for microbial systems and synthetic biology research. They allow tight control of cell culture parameters over long durations. These unique features enable researchers to perform sophisticated experiments and to achieve high experimental reproducibility. A weakness of existing small-scale, low-cost bioreactors is their limited automated measurement capabilities. Researchers usually need to manually extract and process culture samples to run them through specialized instruments. This is usually tedious and strongly constrains the available temporal resolution and scope. It also precludes the dynamic adaptation of culture conditions in response to such measurements.
ReacSight is a generic and flexible strategy to enhance multi-bioreactor platforms for automated measurements and reactive experiment control. On the hardware side, ReacSight leverages a pipetting robot to create a physical link between any multi-bioreactor setup and the input of any plate-based measurement device. On the software side, ReacSight enables full platform integration via a versatile instrument control architecture based on python and the python web application framework Flask. ReacSight software also provides a generic event system to enable reactive experiment control.
6.3 Programmatic control of plate readers with PlateRider
Participants: Virgile Andréani (main developper).
Multiwell plates are simply flat plastic plates having wells that can be used as small test tubes. Plate readers are affordable pieces of equipment that allow to grow cell cultures inside wells, to measure various quantities of interest, such as turbidity or fluorescence, and to dispense inside wells small volumes of reagents.
Because multiwell plates typically contain 96, 384 or even 1536 wells, this simple technology allows to significantly increase the experimental throughput. Yet, experiments have to be manually specified using the graphical user interface of the vendor software by moving blocks of wells. Specifying complex experiments is time-consuming and error-prone.
To circumvent this limitation, we have implemented PlateRider, a driver and an API for Tecan Spark machines, a popular type of plate readers. The software is in Python and consists approximately of a thousand significant lines of code. It supports arbitrarily complex protocols, including reactive experiments, and possess an extensive logging system.
7 New results
In addition to the publications or preprints described below, we would like to mention the results described in the PhD thesis of Virgile Andréani as a significant contribution to the field 1.
7.1 To quarantine, or not to quarantine: A theoretical framework for disease control via contact tracing
Participants: Davin Lunz, Gregory Batt, Jakob Ruess.
Contact tracing via smartphone applications is expected to be of major importance for maintaining control of the COVID-19 pandemic. However, viable deployment demands a minimal quarantine burden on the general public. That is, consideration must be given to unnecessary quarantining imposed by a contact tracing policy. Previous studies have modeled the role of contact tracing, but have not addressed how to balance these two competing needs. In 5, we proposed a modeling framework that captures contact heterogeneity. This allows contact prioritization: contacts are only notified if they were acutely exposed to individuals who eventually tested positive. The framework thus allows us to address the delicate balance of preventing disease spread while minimizing the social and economic burdens of quarantine. This optimal contact tracing strategy was studied as a function of limitations in testing resources, partial technology adoption, and other intervention methods such as social distancing and lockdown measures. The framework is globally applicable, as the distribution describing contact heterogeneity is directly adaptable to any digital tracing implementation.
7.2 Beyond the chemical master equation: stochastic chemical kinetics coupled with auxiliary processes
Participants: Davin Lunz, Gregory Batt, Jakob Ruess.
The chemical master equation and its continuum approximations are indispensable tools in the modeling of chemical reaction networks. These are routinely used to capture complex nonlinear phenomena such as multimodality as well as transient events such as first-passage times, that accurately characterise a plethora of biological and chemical processes. However, some mechanisms, such as heterogeneous cellular growth or phenotypic selection at the population level, cannot be represented by the master equation and thus have been tackled separately. In 4, we developed a unifying framework that augments the chemical master equation to capture such auxiliary dynamics, and we developed and analysed a numerical solver that accurately simulates the system dynamics. We showcased these contributions by casting a diverse array of examples from the literature within this framework, and applied the solver to both match and extend previous studies. Analytical calculations were performed for each example to validate the numerical results and to benchmark the solver implementation.
7.3 On continuum approximations of discrete-state Markov processes of large system size
Participants: Davin Lunz.
Discrete-state continuous-time Markov processes are an important class of models employed broadly across the sciences. When the system size becomes large, standard approaches can become intractable to exact solution and numerical simulation. Approximations posed on a continuous state space are often more tractable and are presumed to converge in the limit as the system size tends to infinity. For example, an expansion of the master equation truncated at second order yields the Fokker–Planck equation, a widely used continuum approximation equipped with an underlying process of continuous state. Surprisingly, in [Doering et. al. Multiscale Model. Sim. 2005 3:2, p.283–299] it is shown that the Fokker–Planck approximation may exhibit exponentially large errors, even in the infinite system-size limit. Crucially, the source of this inaccuracy has not been addressed. In 9, we focused on the family of continuous-state approximations obtained by arbitrary-order truncations. We uncovered how the exponentially large error stems from the truncation by quantifying the rapid error decay with increasing truncation order. Furthermore, we explained why this discrepancy only comes to light in a subset of problems. The approximations produced by finite truncation beyond second order lack underlying stochastic processes. Nevertheless, they retain valuable information that explains the previously observed discrepancy by bridging the gap between the continuous and discrete processes. The insight conferred by this broader notion of “continuum approximation”, where we do not require an underlying stochastic process, prompted us to revisit previously expressed doubts regarding continuum approximations. In establishing the utility of higher-order truncations, this approach also contributed to the extensive discussion in the literature regarding the second-order truncation: while recognising the appealing features of an associated stochastic process, in certain cases it may be advantageous to dispense of the process in exchange for the increased approximation accuracy guaranteed by higher-order truncations.
7.4 Enhancing multi-bioreactor platforms for automated measurements and reactive experiment control
Participants: François Bertaux, Sebastián Sosa Carrillo, Achille Fraisse, Chetan Aditya, Gregory Batt.
New small-scale, low-cost bioreactor designs provide researchers with exquisite control of environmental parameters of microbial cultures over long durations, allowing them to perform sophisticated, high-quality experiments that are particularly useful in systems biology, synthetic biology and bioengineering. However, existing setups are limited in their automated measurement capabilities, primarily because sensitive and specific measurements require bulky, expensive, stand-alone instruments (for example, most single-cell resolved measurements require a cytometer or a microscope). In 2, we present ReacSight, a generic and flexible strategy to enhance multi-bioreactor platforms for automated measurements and reactive experiment control. We use ReacSight to assemble a platform for single-cell resolved characterization and reactive optogenetic control of parallel yeast continuous cultures. We demonstrate its usefulness by achieving parallel real-time control of gene expression with light in different bioreactors and by exploring the relationship between fitness, nutrient scarcity and cellular stress density using highly-controlled and informative competition assays.
7.5 A bacterial size law revealed by a coarse-grained model of cell physiology
Participants: François Bertaux.
Universal observations in Biology are sometimes described as “laws”. In E. coli, experimental studies performed over the past six decades have revealed major growth laws relating ribosomal mass fraction and cell size to the growth rate. Because they formalize complex emerging principles in biology, growth laws have been instrumental in shaping our understanding of bacterial physiology. In 3, we discovered a novel size law that connects cell size to the inverse of the metabolic proteome mass fraction and the active fraction of ribosomes. We used a simple whole-cell coarse-grained model of cell physiology that combines the proteome allocation theory and the structural model of cell division. This integrated model captures all available experimental data connecting the cell proteome composition, ribosome activity, division size and growth rate in response to nutrient quality, antibiotic treatment and increased protein burden. Finally, a stochastic extension of the model explains non-trivial correlations observed in single cell experiments including the adder principle. This work provides a simple and robust theoretical framework for studying the fundamental principles of cell size determination in unicellular organisms.
7.6 Ratiometric quorum sensing governs the trade-off between bacterial vertical and horizontal antibiotic resistance propagation
Participants: Arthur Carcano.
Plasmid-mediated horizontal gene transfer of antibiotic resistance and virulence in pathogenic bacteria underlies a major public health issue. Understanding how, in the absence of antibiotic-mediated selection, plasmid-bearing cells avoid being outnumbered by plasmid-free cells is key to developing counterstrategies. In 6, we quantified the induction of the plasmidial sex pheromone pathway of Enterococcus faecalis to show that the integration of the stimulatory (mate-sensing) and inhibitory (self-sensing) signaling modules from the pCF10 conjugative plasmid provides a precise measure of the recipient-to-donor ratio, agnostic to variations in population size. Such ratiometric control of conjugation favors vertical plasmid transfer under low mating likelihood and allows activation of conjugation functions only under high mating likelihood. We further showed that this strategy constitutes a cost-effective investment into mating effort because overstimulation produces unproductive self-aggregation and growth rate reduction. A mathematical model suggests that ratiometric control of conjugation increases plasmid fitness and predicts a robust long-term, stable coexistence of donors and recipients. Our results demonstrated how population-level parameters can control transfer of antibiotic resistance in bacteria, opening the door for biotic control strategies.
7.7 Optimal design of single-cell experiments within temporally fluctuating environments
Participants: Zachary Fox.
Modern biological experiments are becoming increasingly complex, and designing these experiments to yield the greatest possible quantitative insight is an open challenge. Increasingly, computational models of complex stochastic biological systems are being used to understand and predict biological behaviors or to infer biological parameters. Such quantitative analyses can also help to improve experiment designs for particular goals, such as to learn more about specific model mechanisms or to reduce prediction errors in certain situations. A classic approach to experiment design is to use the Fisher information matrix (FIM), which quantifies the expected information a particular experiment will reveal about model parameters. The finite state projection-based FIM (FSP-FIM) was recently developed to compute the FIM for discrete stochastic gene regulatory systems, whose complex response distributions do not satisfy standard assumptions of Gaussian variations. In 7, we develop the FSP-FIM analysis for a stochastic model of stress response genes in S. cerevisiae under time-varying MAPK induction. We verify this FSP-FIM analysis and use it to optimize the number of cells that should be quantified at particular times to learn as much as possible about the model parameters. We then extend the FSP-FIM approach to explore how different measurement times or genetic modifications help to minimize uncertainty in the sensing of extracellular environments, and we experimentally validate the FSP-FIM to rank single-cell experiments for their abilities to minimize estimation uncertainty of NaCl concentrations during yeast osmotic shock. This work demonstrates the potential of quantitative models to not only make sense of modern biological datasets but to close the loop between quantitative modeling and experimental data collection.
7.8 Diverse cell stimulation kinetics identify predictive signal transduction models
Participants: Zachary Fox.
Computationally understanding the molecular mechanisms that give rise to cell signaling responses upon different environmental, chemical, and genetic perturbations is a long-standing challenge that requires models that fit and predictquantitative responses for new biological conditions. Overcoming this challenge depends not only on good models and detailed experimental data but also on the rigorous integration of both. In 8, we propose a quantitative framework to perturb and model generic signaling networks using multiple and diverse changing environments (hereafter ‘‘kinetic stimulations’’) resulting in distinct pathway activation dynamics. We demonstrate that utilizing multiple diverse kinetic stimulations better constrains model parameters and enables predictions of signaling dynamics that would be impossible using traditional dose-response or individual kinetic stimulations. To demonstrate our approach, we use experimentally identified models to predict signaling dynamics in normal, mutated, and drug-treated conditions upon multitudes of kinetic stimulations and quantify which proteinsand reaction rates are most sensitive to which extracellular stimulations.
8 Partnerships and cooperations
8.1 International initiatives
-
ANR-FWF CyberCircuits (2018-2022), on “Cybergenetic circuits to test composability of gene networks”, co-coordinated by C. Guet (IST Austria, Austria) and Jakob Ruess.
The objective of the Cybercircuit project is to explain and better predict how composed circuits function in vivo. To tackle this long standing question, we will construct hybrid bio-digital circuits in which a part of a network is effectively implemented as a biological genetic network whereas another part exists only virtually in the form of a model in a computer.
8.2 International research visitors
Lorenzo Pasotti, assistant professor at the Department of Electrical, Computer and Biomedical Engineering and at the Centre for Health Technologies of the University of Pavia (Italy) has been invited for several month in the InBio team.
8.3 European initiatives
8.3.1 FP7 & H2020 Projects
-
FET Open COSY-BIO (2017-2021), on “Control Engineering of Biological Systems for Reliable Synthetic Biology Applications, coordinated by Diego di Bernardo (Tigem), with Filippo Menolascina (Edinburgh U), Mario di Bernardo (Naples U) , Pascal Hersen (Paris7 U), Mustafa Khammash (ETHZ), Grégory Batt, Guy-Bart Stan (Imperial College), and Lucia Marucci (Bristol U).
The main objective of COSY-BIO is to identify generally applicable approaches to design closed-loop feedback controllers for biological systems. The project will rely on control engineering for physical systems and on the exploitation of the unique features of living organisms.
8.4 National initiatives
8.4.1 ANR Projects
-
ANR MEMIP (2016-2021) on “Mixed-Effects Models of Intracellular Processes”, coordinated by G. Batt, with P. Hersen, (CNRS/Paris7), E. Cinquemani (IBIS, Inria) and M. Lavielle (XPOP, Inria/CNRS/Polytechnique).
The objective of the MEMIP project is to develop mathematical methods for the identification of single-cell models, and to implement and bendchmark them on experimental data generated by innovative experimental platforms.
-
ANR CoGEx (2016-2020) on “Computer Aided Control of Gene Expression”, coordinated by P. Hersen (MSC lab, CNRS/Paris7), with G. Batt and G. Truan (LISBP, CNRS/INSA);
The CoGEx project aims at developing experimental and theoretical tools for the computer-based remote-control of live cells, and to use such a system to interrogate cellular processes at the single cell level.
-
Institut de Convergence Inception (2016-2025) on the “Emergence of Diseases in Populations and in Individuals”, coordinated by T. Bourgeron (Institut Pasteur). Partner institutes include Institut Pasteur, Paris Sciences et Lettres, Université de Paris, AP-HP, and research teams from CEA, CNRS, INSERM and INRA.
The Inception’s goal is to develop a core structure to mobilize data resources, numerical sciences, and fundamental experimental biology in a range of health issues. It uses integrative biology, social science and data science to understand the emergence of diseases in populations and in individuals.
8.4.2 Inria Project Labs
-
IPL COSY (2017-2021) on “Real-time control of synthetic microbial communities”, coordinated by Eugenio Cinquemani (Ibis, Inria), with Jean-Luc Gouzé (Biocore, Inria), Grégory Batt, Frédéric Bonnans (Commands, Inria), Efimov Denis (Non-A, Inria), and Hans Geiselmann (BIOP, Université Grenoble-Alpes), Béatrice Laroche (Maiage, Inra Jouy-en-Josas).
The COSY project aims at developping automated experimental platforms and control methods for heterogeneous microbial populations to propose solutions for the optimization of bioproduction processes.
9 Dissemination
9.1 Promoting scientific activities
9.1.1 Scientific events: organisation
In 2020, the working group Biologie Systémique Symbolique (BIOSS) organized a series of monthly seminars in lieu of its usual yearly working days. In 2020, Gregory Batt, a member of the steering committee, helped with their organizations.
9.1.2 Scientific events: selection
Member of the conference program committees
- Jakob Ruess was a member of the program committee of Bioinformatics 2020, the 11th International Conference on Bioinformatics Models, Methods and Algorithms. Valetta, Malta, February 2020.
Reviewer
- Jakob Ruess was a reviewer for Bioinformatics 2020 and for the European Control Conference (ECC 2021).
9.1.3 Journal
- François Bertaux has been a reviewer for Bioinformatics
- Gregory Batt has been a reviewer for Nature Communications.
- Jakob Ruess has been a reviewer for IEEE Transactions on Automatic Control, mSystems, The FEBS Journal, Theoretical Computer Science and Frontiers Public Health journals.
9.1.4 Invited talks
- Grégory Batt gave a keynote adress at the 18th conference on Computational Methods in Systems Biology (CMSB 2020) in September 2020. He also gave an invited seminar at the Infection, Antimicrobials, Modelling, and Evolution research center in Paris in September 2020.
- Olivier Borkowski has been an invited speaker at the 5th Applied Synthetic Biology in Europe conference in Delft, Netherland, in November 2020. He has also been an invited speaker at the Beckman AIChE webminar.
- Davin Lunz gave an invited presentation at the seminar of the Modélisation et Covid-19 working group in December 2020.
- Virgile Andréani gave an invited presentation at the seminar of the Genomes and Genetics department of Institut Pasteur in February 2020
9.1.5 Leadership within the scientific community
Grégory Batt is member of the Technical Committee on Systems Biology of IEEE and CSS societies.
He is also member of the scientific board of the French research network on Bioinformatics (GdR BIM) and a member of the steering committee of the working group on Symbolic Systems Biology (GT Bioss), affiliated to two French research networks, on Bioinformatics (GdR BIM) and on Mathematical computer science (GdR IM).
9.1.6 Scientific expertise
Grégory Batt has been a member of the strategic and scientific committee of the Grand Défi initiative on biomedical drugs and bioproduction of the French Conseil de l'Innovation and operated by the ANR and Bpifrance. He has also been an expert to Bpifrance, the French public investment bank, for the evaluation of Projets Structurants Pour la Compétitivité. Lastly he has also provided expertise to asses a joint research initiative between the Sup'BioTech and the IPSA enginering schools.
At Institut Pasteur, Francois Bertaux has been member of the Elab working group, a significant initiative for the deployment of electronic lab notebooks throughout the institute. Gregory Batt has been member of the working group on data management and scientific reproducibility.
9.1.7 Research administration
Grégory Batt is deputy director of the Computational Biology Department at Institut Pasteur. Department directors have an important advisory role to the direction and are involved in hiring processes. He has also being director of the USR 3756 CNRS unit since August 2020, and interim department director directeur in August and September 2020.
9.2 Teaching - Supervision - Juries
9.2.1 Teaching
- Grégory Batt (42h), Jakob Ruess (28h), and Arthur Carcano (28h), Computational Biology, M1, Master Interdisciplinary Approaches to Life Sciences (AIRE-LiSc).
- François Bertaux (12h), Systems Biology, M1, Master Interdisciplinary Approaches to Life Sciences (AIRE-LiSc).
- Olivier Borkowski (5h), Synthetic Biology, M2 Master AgroParisTech.
9.2.2 Supervision
- PhD defended: Virgile Andréani, “Modelling and efficient characterization of enzyme-mediated response to antibiotic treatments“, ED IPP, Ecole Polytechnique, Defended on Dec 16, 2020. Supervision by Grégory Batt.
- PhD in progress: Chetan Aditya, “Control of heterogenous synthetic microbial systems”, ED FdV, Université de Paris, Started in Feb. 2018. Supervision by Grégory Batt and Jakob Ruess.
- PhD in progress: Arthur Carcano, “Iterative design of single-cell experiments to learn single-cell models of biological systems”, ED FIRE, Université de Paris. Started in Oct. 2018. Supervision by Grégory Batt.
- PhD in progress: Sebastian Sosa Carrillo, “Understanding the cost of protein production in yeast”, ED FIRE, Université de Paris. Started in Feb. 2018. Supervision by Grégory Batt and François Bertaux.
- PhD in progress: Viktoriia Gross, “An integrative approach to characterize the two sides of enzyme-mediated antibiotic escape: resistance and tolerance. Started in Oct. 2020. Supervision by Imane El Meouche, Erick Denamur and Grégory Batt.
9.2.3 Juries
Olivier Borkowski has been a reviewer of the PhD thesis of Dominic Logel at Macquarie University, Australia, and a member of the jury for his PhD defense in July 2020.
10 Scientific production
10.1 Major publications
- 1 phdthesis Modelling and Efficient Characterization of Enzyme-Mediated Response to Antibiotic Treatments Ecole polytechnique December 2020
- 2 unpublishedEnhancing multi-bioreactor platforms for automated measurements and reactive experiment controlDecember 2020, working paper or preprint
- 3 article A bacterial size law revealed by a coarse-grained model of cell physiology PLoS Computational Biology 16 9 September 2020
- 4 unpublishedBeyond the chemical master equation: stochastic chemical kinetics coupled with auxiliary processesNovember 2020, working paper or preprint
- 5 article To quarantine, or not to quarantine: A theoretical framework for disease control via contact tracing Epidemics 34 December 2020
10.2 Publications of the year
International journals
Doctoral dissertations and habilitation theses
Reports & preprints

