Keywords
Computer Science and Digital Science
- A3.1.1. Modeling, representation
- A3.1.10. Heterogeneous data
- A3.1.11. Structured data
- A3.3.2. Data mining
- A3.3.3. Big data analysis
- A3.4.1. Supervised learning
- A3.4.2. Unsupervised learning
- A3.4.3. Reinforcement learning
- A3.4.4. Optimization and learning
- A3.4.5. Bayesian methods
- A5.2. Data visualization
- A6.1.1. Continuous Modeling (PDE, ODE)
- A6.2.4. Statistical methods
- A6.3.1. Inverse problems
- A6.3.4. Model reduction
- A6.4.2. Stochastic control
- A9.2. Machine learning
Other Research Topics and Application Domains
- B1.1. Biology
- B1.1.5. Immunology
- B1.1.7. Bioinformatics
- B1.1.10. Systems and synthetic biology
- B2.2.4. Infectious diseases, Virology
- B2.2.5. Immune system diseases
- B2.3. Epidemiology
- B2.4.1. Pharmaco kinetics and dynamics
- B2.4.2. Drug resistance
- B9.5.6. Data science
- B9.8. Reproducibility
1 Team members, visitors, external collaborators
Research Scientists
- Quentin Clairon [INRIA, Researcher, from Oct 2022]
- Boris Hejblum [INSERM, Researcher]
- Melanie Prague [INRIA, Researcher]
Faculty Members
- Rodolphe Thiebaut [Team leader, UNIV BORDEAUX and UNIV HOSPITAL BORDEAUX, Professor, HDR]
- Marta Avalos Fernandez [UNIV BORDEAUX, Associate Professor, HDR]
- Robin Genuer [UNIV BORDEAUX, Associate Professor, HDR]
- Edouard Lhomme [UNIV BORDEAUX and UNIV HOSPITAL BORDEAUX, Associate Professor]
- Laura Richert [UNIV BORDEAUX and UNIV HOSPITAL BORDEAUX, Professor, HDR]
- Linda Wittkop [UNIV BORDEAUX and UNIV HOSPITAL BORDEAUX, Associate Professor, HDR]
Post-Doctoral Fellows
- Marie Alexandre [INRIA, from Jun 2022]
- Mathieu Chalouni [ANRS]
- Quentin Clairon [INRIA, until Sep 2022]
PhD Students
- Marie Alexandre [INRIA, until May 2022]
- Kalidou Ba [INSERM, from Sep 2022]
- Houreratou Barry [ Institut National de Santé de Publique du Burkina Faso]
- Thomas Ferte [UNIV HOSPITAL BORDEAUX]
- Iris Ganser [INSERM]
- Benjamin Hivert [INSERM]
- Hélène Savel [IPSEN]
Technical Staff
- Kalidou Ba [INSERM, Engineer, until Sep 2022]
- Antonin Colajanni [INSERM, Engineer, from Dec 2022]
- Mélanie Huchon [INSERM, Engineer]
- Clément Nerestan [INRIA, Engineer]
- Anton Ottavi [INSERM, Engineer]
- Panthea Tzourio [INSERM, Engineer]
Interns and Apprentices
- Vishwa Elankumaran [INRIA, Intern, from Mar 2022 until Aug 2022]
- Raphaël Garcia [INSERM, Intern, from Apr 2022]
- Marie Poupelin [INSERM, Intern, from Apr 2022 until Jul 2022]
- Justine Remiat [INSERM, Intern, from Apr 2022 until Jun 2022]
- Florian Robert [INRIA, Intern, from Jun 2022 until Sep 2022]
- Bhushan Rugbursing [INSERM, from May 2022 until Jul 2022]
- Malik Sari [INSERM, Intern, from Jul 2022 until Sep 2022]
- Simon Valayer [INRIA, Intern, from Apr 2022]
Administrative Assistants
- Sandrine Darmigny [INSERM]
- Audrey Plaza [INRIA, until Sep 2022]
- Rima Soueidan [INRIA, from Sep 2022]
Visiting Scientists
- Daniel Dori [ Université Joseph Ki-Zerbo, Burkina Faso, from Jun 2022 until Jun 2022]
- Baptiste Elie [Ecole Normale Supérieure Paris-Saclay, from Jun 2022 until Jun 2022, PhD student with Samuel Alizon, College de France. Travel grant / scientific exchange ANRS MIE]
- Jane Heffernan [York University of Toronto , from May 2022 until May 2022, Professor of mathematical modelling ]
- Isabela Ornelas [Brasilian Ministry of Health (Brasil), from 17 October to 15 December 2022 , from Oct 2022, Epidemiologist]
2 Overall objectives
The two main objectives of the SISTM team are:
-
i)
to accelerate the development of vaccines by analyzing all the information available in early clinical trials and optimizing new trials
-
ii)
to develop new data science approaches to analyze and model high dimensional data in small sample size studies.
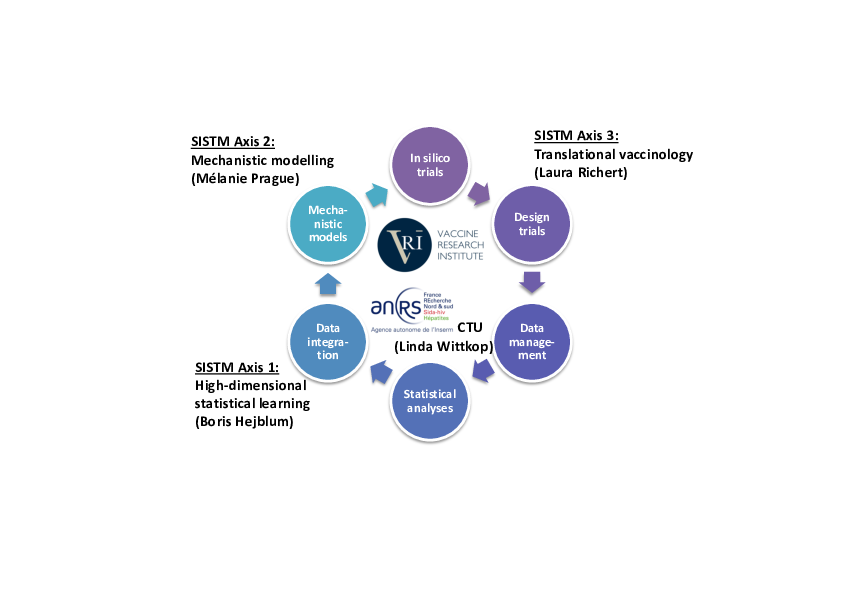
The methods developed are relevant in many other applications than those encountered in the SISTM team. However, the focus devoted to vaccine development is justified by the importance of the objective from a public health point of view and a good knowledge of the application field that maximizes the relevance and the implementation of the methods developed. This equilibrium between the methodological and the applied work reached over the last years is a fundamental motivation for each member of the SISTM team even though the background could be very different from one researcher to the other (e.g. applied mathematics vs. public health). This equilibrium is maintained by the organization of the team as well as the collaborations established especially through the Vaccine Research Institute, Bordeaux University, Inserm and Inria. Hence, we are able to collaborate for a theoretical problem during the development of a new method (e.g. demonstration of the convergence of an estimator) as well as to translate the research outcomes (either new analytical methods or applied results) to clinicians and biologists first in our collaborative networks and then beyond. Figure (1) illustrates this synergism and materializes the three research axis of the team: high dimension statistical learning, mechanistic modelling and translational vaccinology.
Biological and clinical research has dramatically changed thanks to technological advances, leading to the possibility of measuring much more biological parameter in high-throughput methods than previously. Clinical research studies can include now traditional measurements such as clinical status, but also (tens of) thousands of cell populations, peptides, gene expressions for a given participant. This has facilitated the transfer of knowledge from basic to clinical science (from ”bench to bedside”) and vice versa, a process often called “Translational medicine”. However, the analysis of these large amounts of data requires specific methods, especially when one wants to have a global understanding of the information inherent to complex systems through an “integrative analysis”. Systems like the immune system are complex because of the many interactions within and between several levels (inside cells, between cells, in different tissues, between individuals, between various species). This has led to a new field called “Systems biology” rapidly adapted to specific topics such as “Systems Immunology” 73, “Systems vaccinology” 71, “Systems medicine” 60. From the statistical point of view, two main challenges arise: i) to adequately deal with the massive amount of data ii) to find relevant models capturing observed data.
First, with respect to the relatively moderate number of participants in vaccine studies and clinical trials, this profusion of high-throughput “omics” data often sets us in a ultra high-dimension context. This mandates updated statistical tools able to tackle this wealth of information. On top of the challenge signal extraction and dimension reduction, there is a redundancy of the information across data modalities, that in turn can be leveraged to boost statistical methods and harness artificial intelligence approaches to predict immunological surrogate endpoints from early indicators.
Second, once a small amount of markers has been selected, we use modeling approaches to understand the biological mechanism (specifically in vaccinology antibodies kinetics or viral dynamics 67). In our work we are interested in the inverse problem: how can we infer the mechanism of a biological process from data. It can be modeled using differential equations (mainly ordinary but could extend to partial and stochastic). The challenge in our methods rely in the type of collected data which are sparse (as opposed to measured in continuous time), with measurement error and repeated across multiple individuals. Thus, we adopt nonlinear mixed-effects model population approach 64. Construction of these models is a challenging process which requires confirmed expertise, advanced statistical methods and the development of software tools.
Finally, once a model has been defined and validated, it is possible to perform in silico trials to predict further strategies. In particular, a systems personalized vaccinology approach 68 using multidimensional immunogenicity data from clinical trials and statistical models (such as optimal control or reinforcement learning) can help improve the selection of optimized vaccine strategies that can then be tested again in subsequent clinical trials.
Domains of application of our methods in vaccinology focuses on, but not limited to, Ebola virus, Human Immunodeficiency Virus (HIV) virus and SARS-CoV-2 virus. The choice of these applications is
deliberate and important for the relevance of the results and their translation into practice, thanks to a longstanding collaboration with several immunology research teams and the implication of the team in
The SISTM team benefits from a very rich ecosystem. Firstly, it is one of the rare (n6 as of 01/2022) teams belonging to both Inserm and Inria national institutes, which helps establishing collaboration as testified by the co-supervision of PhD Students and co-publications with other researchers belonging to Inserm teams or Inria teams from the two research centres in Bordeaux. Secondly, the applications in clinical research are facilitated by the very close collaboration with Clinical Trial Units (CTUs): from the ANRS/VRI (CMG directed by LW), from Bordeaux Hospital (USMR directed by LR and previously by RT), from F-CRIN (Euclid platform, directed by LR and EL), from the international consortia linked to the Vaccine Research Institute (for which SISTM is leading the data science division). Finally, the team is very much involved in teaching activities at Bordeaux University and ISPED Institute, especially through the Graduate’s program Digital Public Health (directed by RT) and the Master of Public Health (first year in e-learning led by MA, Biostatistics led by RG and Public Health Data Science led by RT). A better description of all these interaction can be found in section Teaching (10.2) and section Fundings (9).
In term of positioning in regards of other teams at Inria and in France, the application domain (immunology and vaccine development) is nearly unique with the exception of DRACULA in Lyon. DRACULA like other teams at Inria (MONC, CARMEN, M3DISIM) or Inserm (IAME) or international groups (e.g. A. Perelson lab in Los Alamos) are also developing mathematical models but rarely with the integration of high dimensional data. In other hand, groups such as Raphael Gottardo lab in Lausanne (previously at the Fred Hutchinson in Seattle) are developing methods for high dimensional data in immunology but are not using dynamical models.
3 Research program
The team is organized in three research axes:
1. High Dimensional Statistical Learning (leader Boris Hejblum),
2. Mechanistic learning (leader Mélanie Prague),
3. Translational vaccinology (leader Laura Richert).
3.1 Axis 1 - High-Dimensional Statistical Learning
The specific objectives are:
- To unlock the analysis of high-dimensional longitudinal data by developing suitable statistical approaches, in particular for applications to high-throughput data (e.g. microbiome, transcriptome, cytomics) generated in vaccine trials.
- To leverage prior biological knowledge and formally incorporate it into statistical models to tackle the small large setting, one of the characteristics of early phase vaccine trials.
- To advance adaptive clustering methods of high-dimensional data in both supervised and unsupervised settings, especially to infer the proportions of cellular population from gene expression measurements and also to identify gene whose expression is key in segmenting transcriptomic measurements across vaccine arms or disease severity for instance.
- To perform feature selection and dimension reduction of high-dimensional molecular and cellular data, as a first step to feed such information into mechanistic models.
Despite being high-dimensional, biomedical data from high-throughput technologies is rarely analyzed in its entirety due to its size or its complexity. For example, in cellular phenotyping data, only a limited number of markers are used to quantify a pre-defined set of cell types; this strategy precludes the discovery of new cell types defined by new combinations of markers. This issue is exacerbated by mass cytometry technologies, which enable the measurement of up to 100 markers on a single cell.
However, measuring specific cells across a large number of intracellular and surface markers requires substantial amounts of blood, ideally fresh, making it difficult to implement such measurements on large sample sizes with multiple repeated measurements. This motivates the exploration of replacing cell phenotyping with transcriptomics analysis in whole blood, as gene expression can be measured more easily and frequently with a much finer temporal resolution (using finger prick at-home self-sampling technology 76). This ambitious endeavor goes beyond previous work done on this topic using standard deconvolution approaches 62. By using more sophisticated statistical 53, machine learning 54, and artificial intelligence 78 models (in particular for adaptive clustering, robust to unobserved cell populations), by exploiting public databases of cytometry data coupled newly available single cell transcriptomics measurements, and by explicitly leveraging the repeated aspect of longitudinal observations from vaccine trial measurements, we set ourselves to successfully study and develop methods delivering accurate cell proportions estimates from gene expression data.
In addition, among high-throughput omics data, the microbiome is also becoming an increasingly important component in understanding the immune system 65. The compositional nature of these data, along with their hierarchical phylogenic structure particularly suited to tree-based models, coupled with their high-dimension requires the use of adequate statistical tools 75.
Furthermore, while those high-throughput molecular and cellular data have an unquestionable value for diving into underlying mechanisms governing and deepening our understanding of the human immune system, we want to determine whether they could be used as early surrogate markers for correlates of protection in vaccine studies (such as antibody titers after vaccination). Due to their high-dimensional nature, answering this question requires the development of new mediation approaches 12 to develop this emerging field of vaccinomics epidemiology.
Outside biological data generated in clinical trials, electronic health records from hospital data warehouse systems are also representing an opportunity for studying infectious diseases and requires specific approaches. Several works have been done on this topic in the SISTM team 57, 59, 74, 81, 22.
Regarding this research axis, there are some common interest with other Inria teams such as HeKA and PreMeDICaL in regards of the use of machine learning approaches applied to medical data or Soda, Mind and Aramis that are more focus on brain applications. Applications in SISTM are focused on analyzing high-throughput omics data (nearly no imaging) in immunology and vaccine trials. Also, modeling biological networks as done in Beagle or Dyliss Inria teams is not an objective of SISTM, the data recorded in human clinical trials being unsuited because of their sparsity. At the international level, the main competitors are groups engaged in biostatistical methods development for the analysis of omics data such as Jeff Leek (previously at John Hopkins, now at Fred Hutchinson, Seattle), Raphael Gottardo (previously at Fred Hutchinson, Seattle, now at Université de Lausanne, Switzerland) or Mark Robinson (Prof at the University of Zurich, Switzerland).
3.2 Axis 2 - Mechanistic learning
The specific objectives are:
- To develop methods for statistical inference of differential equations model parameters in population framework.
- Modeling immunological and virological dynamics in population.
- Implementing control strategies toward personalized medicine.
When studying the dynamics of some given markers one can for instance use descriptive models summarizing the dynamics over time in term of slopes of the trajectories 77. These slopes can be compared between treatment groups or according to patients’ characteristics. Mechanistic modeling, that is dynamical models based on Ordinary Differential Equations (ODE), could be preferred as it integrates knowledge about the biological mechanism and it carries causal interpretation of the observed phenomenon 52, 69. Thus, in this axis, we focus on inference of model parameters of mechanistic models in population of subjects (e.g. from a clinical trial). This modeling is constituted by three features: 1/ a dynamical model, which describes a phenomenon, often based on ODE (but also possibly partial and stochastic DE) 2/ a statistical model, which describes the variability that exists in data, and 3/ an observational model, which relates what is observable with error in the mathematical model.
The definition of the model needs to identify the parameter values that fit the data. Contrary to Inria team such as MAKUTU or BEAGLE, which are interested in simulation scheme for large differential equation systems, we focus on inverse problems for inference of parameters from data. In clinical research, this is challenging because data are sparse, and often unbalanced, coming from populations of individuals. A substantial inter-individual variability is always present and needs to be accounted as this is the main source of information. Many approaches have been developed to estimate the parameters of non-linear mixed models (NLME) including Bayesian approach 80, semi-parametric approaches 79 or penalized likelihood approach (in house NIMROD program 70). The SAEM algorithm 63, as implemented in Monolix 72, is now also used for many of our projects. We however, continue to participate in the development of related methods in collaboration with Inria team XPOP. We also devote a large part of this axis methodological research to the development of alternative methods for estimation in NLME ODE models.
Having a good mechanistic model with a population approach in a biomedical context opens doors to various applications beyond a good understanding of the data. Global and individual predictions can be excellent because of the external validity of a model based on biological mechanisms rather than simple regressions. Control theory (Inria team ASTRAL), game theory (Inria team SCOOL) and learning approaches (Inria team FLOWERS) may serve for defining optimal interventions or optimal designs to evaluate new interventions. In the last period of evaluation, we made a proof of concept of such open-loop control problem. We model the response to Interleukin-7 (IL-7) injections in HIV-infected patients, and that has allowed to design new trials finally implementing personalized medicine 61. We still devote a large part of this axis methodological research to the development of methods around personalized medicine.
Finally, one core research direction of this axis, is the development of statistically proven (unbiased and efficient) methods for evaluation of intervention effects. Once adjusted to data, these models could be used to perform in silico trials to predict the effect of a various administration strategies in various populations. Development of estimation/simulation tools allow the translational vaccinology axis to better design next trials.
Regarding this axis, the SISTM team compares to DRACULA, BIOCORE, MONC and COMPO Inria team. However, differences arise in two ways 1/ the application field is immunology and infectious diseases and 2/ we adopt a population approach. This last point results in using simpler models in which it is possible to infer parameters from sparse data by taking advantage of an underlying mechanism common to all patients.
3.3 Axis 3 - Translational vaccinology
The specific objectives are:
- To accelerate the vaccine development by in depth analysis of data generated in early clinical trials and
- designing the next trials in collaboration with immunologists and clinicians.
Vaccines are one of the most efficient tools to prevent and control infectious diseases, and there is a need to increase the number of safe and efficacious vaccines against various pathogens. However, clinical development of vaccines - and of any other investigational product - is a lengthy and costly process. Considering the public health benefits of vaccines, their development needs to be supported and accelerated. During early phase clinical vaccine development (phase I, II, translational trials), the number of possible candidate vaccine strategies against a given pathogen that needs to be down-selected is potentially very large. Moreover, during early clinical development there are most often no validated surrogate endpoints to predict the clinical efficacy of a vaccine strategy based on immunogenicity results that could be used as a consensus immunogenicity endpoint and down-selection criterion. This implies considerable uncertainty about the interpretation of immunogenicity results and about the potential value of a vaccine strategy as it transits through early clinical development. Given the complexity of the immune system and the many unknowns in the generation of a protective immune response, early vaccine clinical development nowadays thus takes advantage of high throughput (or “omics”) methods allowing to simultaneously assess a large number of response markers at different levels (“multi-omics”) of the immune system. Outside of the context of emergency vaccine development during a pandemic, this has induced a paradigm shift towards early-stage and translational vaccine clinical trials including fewer participants but with thousands of data points collected on every single individual. This is expected to contribute to acceleration of vaccine development thanks to a broader search for immunogenicity signals and a better understanding of the mechanisms induced by each vaccine strategy. However, this remains a difficult research field, both from the immunological as well as from the statistical perspective. Extracting meaningful information from these multi-omics data and transferring it towards an acceleration of vaccine development requires adequate statistical methods (in close collaboration with axis 1), state-of-the art immunological technologies and expertise, and thoughtful interpretation of the results.
Our main current areas of application here are early phase trials of HIV and Ebola vaccine strategies, in which we participate from the initial trial design to the final data analyses. We are also involved in the development of next-generation pan-Coronavirus vaccines.
In regards of the number of trials we are dealing with, the complexity of the data (including clinical and biological high dimensional data), the need for a collaborative tool for data sharing that is respectful of GDPR and health data protection, we have set up a data warehouse system based on the Labkey solution (also used for the Immunespace funded by the NIH). We are currently plugging in our data analysis and data vizualization tools. This solution may constitute a very nice way to boost our collaborations but also to facilitate the access to the statistical tools we have developped.
To our knowledge, our specific application to vaccine trials is unique in France. Although some research teams have sometimes applications in this field (e.g. clinical epidemiology team at Inserm U1018 or Inria DRACULA team), there are less devoted to it. Internationally, the closest group to SISTM research axis 3 is the vaccine and infectious disease division of the Fred Hutchinson Institute (Seattle). There are also several groups working on systems immunology mainly in United States such as Mark Davis at Stanford University, Bali Pulendran at Emory University, Rafick Sekaly at Case Western Reserve University, Galit Alter at the Ragon Institute. There are all immunologists integrating bioinformaticians in their groups therefore they are more applying than developping new methods. We have collaborated with several of these groups.
4 Application domains
The main application domain is the clinical immunology of infectious diseases and more specifically vaccine development.
The main infectious diseases concerned up to now are:
- Human Immunodeficiency Virus (HIV);
- Ebola virus (following the 2014 epidemics);
- SARS-Cov2 virus;
- Hepatitis B virus;
- NIPAH virus;
This is not a closed list and new studies are currently settled on other infectious agents (e.g. tuberculosis, Human Papilloma Virus...).
5 Social and environmental responsibility
5.1 Footprint of research activities
-
National and international programs
- Coordination of the response to the Referral for primary care clinical research in France - Ministry of Health (September 2021 - April 2022): The objective was to make proposals to anticipate the implementation of future ambulatory trials in response to an emerging infectious disease and enable them to reach their recruitment targets quickly, and to structure research in primary care more broadly. The response includes a national and international review of COVID-19 ambulatory research and 20 proposals on research strategy, its structuring and the removal of budgetary and regulatory constraints.
- Participation in Delphi consensus groups: The objective was to extend the CONSORT recommandations. Participated in the elaboration of SPIRIT/CONSORT Extension for Dose-finding Trials (2022)
5.2 Impact of research results
-
Drug licensure and patents
- Participant as "Inventor" (Décret n°96-858 du 2 octobre 1996) to the development and the authorization for commercialization (1/7/2020) of the Janssen Zabdeno® (Ad26.ZEBOV) and Mvabea® (MVA-BN-Filo) vaccines against Ebola virus infection.
- Patent 20 306 527.1 on "Use of CD177 as biomarker of worsening in patients suffering from COVID-19" (10/12/2020)
-
Public/Private partnership
- In the context of clinical trials: Johnson and Johnson (IMI-2 Anti-Ebola vaccine trial Ebovac and Prevac; Merck (Anti-Ebola vaccine trial Prevac/Prevac-up); Iliad Biotechnologies (Anti-pertussis vaccine trial BPZE-1); Gilead Sciences (IP-Cure-B)
- In the context of CIFRE PhD funding: Ipsen (LR HS, 2020- )
-
Multicenter clinical trials on vaccine research
- Coordination clinical trials through the Euclid/F-CRIN, CIC1401 platform: Leading Phase II international clinical trials (steering and methodology) for projects BPZE-1, Ebovac2, IP-Cure-B, Prevac, Prevac-Up et PrimalVac (see fundings section).
- Methodology for clinical trials:
- International phase II anti-Ebola vaccine trial PREVAC (NCT02876328) and EDCTP2 PREVAC-UP
- International phase I anti-Malaria vaccine trial PRIMALVAC (NCT02658253)
- French Phase I/II anti-HIV vaccine trial ANRS VRI01 (NCT02038842)
- French Phase I anti-HIV vaccine trial ANRS VRI06 (NCT04842682)
- Monocenter anti-pertussis phase I vaccine trial BPZE-1 (NCT02453048)
- French phase II anti-pneumococal vaccine trial PNEUMOVAS (NCT03069703)
- French phase II anti-pneumococal vaccine trial SPLENEVAC2 (NCT03873727)
- French phase II anti-meningococcal vaccine trial SPLENMENGO (NCT04166656)
- French phase II anti-HPV vaccine trial PRIMAVERA (NCT01687192)
- French Phase IV anti-Dengue vaccine trial (LR, trial set-up ongoing)
- Cohort study of anti-COVID-19 vaccination in specific populations (ANRS0001S COV-POPART)
- Cohort study of HIV infected patients in Nouvelle-Aquitaine (ANRS CO3 Aquitaine)
- Cohort study of HIV-2 infected patients in France (ANRS CO5 VIH-2)
- Cohort study of co-infected patients with HIV and Hepatitis in France (ANRS CO13 HEPAVIH )
- International phase II proof of concept trial IP-cure-B . Educating the liver immune environment through TLR8 stimulation followed by NUC discontinuation. (ANRS HB 07 IP-Cure-B Trial)
6 Highlights of the year
The SISTM team has been evaluated by INRIA with motivating feedback from the external reviewers (e.g. "I can only applaud the strong vision and achievements of the team, which is on the path to be one of the world-leading groups in the field.").
The team has been reinforced by Quentin Clairon, recruited as INRIA researcher (IFSP).
A strong effort has been done to settled our data warehouse (based on Labkey solution) to be ready for 2023. It includes all raw data and meta-data on the design of the clinical trials and it is used in international collaborations to facilitate data sharing and exploration (EHVA, EBOVAC, IP-Cure-B and CARE consortia).
In a methodological point of view:
- We contributed to an important work published in Biostatistics in collaboration with our new DESTRIER associate team where we developed a method for the evaluation of surrogate marker in high-dimensional settings.
- We have developed a new mechanistic model for the effect of SARS-Cov2 vaccine in infected macaques quantifying the effect of vaccines on the cell infection rates which has been published in eLife.
In regards of the translation to clinical vaccinology:
- A large randomized multi-arm Ebola vaccine trial (Prevac trial, evaluating three different Ebola vaccine strategies) has had its primarily results available and a manuscript has been published in the New England Journal of Medicine (December 2022).
- The ANRS VRI01 phase I/II vaccine trial, for which we had developed an innovative early phase design has been completed and final results published.
- The ANRS VRI06 first-in-human phase I trial of a novel vaccine concept tar- geting dendritic cells (here as HIV vaccine) has started in 2021 and the fist results will be presented at CROI 2023.
- An important milestone is also the set-up and conduct of the national CovPopArt Covid Vaccine Cohort (co-PI: LW) : a French cohort for assessing COVID-19 vaccine responses in specific populations, with results becoming available in 2022 (Loubet, Wittkop et al. CMI 2022)
- Covid treatment trials: The researchers of the axis have been heavily mobilized by the Coverage France clinical trial. Coverage France is a national multi-arm multi-stage (MAMS) adaptive trial platform for early treatments in Covid-infected outpatients in France that has been set-up in spring 2020 and obtained the “national priority” label (Capnet) [100, 52]. The trial has been adapted several times since its start, including evolution of the design and the evaluation of new treatment strategies, and is stopped since December 2021. A manuscript with the results of one of the tested treatment strategies (ciclesonide) has been published in 2022.
7 New software and platforms
7.1 New software
7.1.1 VALIDICLUST
-
Name:
VALID Inference for Clusters Separation Testing
-
Keywords:
Biostatistics, Statistics, Statistical inference
-
Functional Description:
R package for post-clustering inference. Given a partition resulting from any clustering algorithm, the implemented tests allow valid post-clustering inference by testing if a given variable significantly separates two of the estimated clusters. 3 tests are implemented: test_selective_inference and merge_selective_inference (both using the selective inference framework) and test_multimod (using the concept of multimodality to describe clusters separation)
- URL:
-
Contact:
Benjamin Hivert
7.1.2 CytOpT
-
Name:
Optimal Transport for Gating Transfer in Cytometry Data with Domain Adaptation
-
Keywords:
Python, Single cell, Computational biology, Bioinformatics, Biomarkers, Biostatistics
-
Functional Description:
Supervised learning from a source distribution (with known segmentation into cell sub-populations) to fit a target distribution with unknown segmentation. It relies regularized optimal transport to directly estimate the different cell population proportions from a biological sample characterized with flow cytometry measurements. It is based on the regularized Wasserstein metric to compare cytometry measurements from different samples, thus accounting for possible mis-alignment of a given cell population across sample (due to technical variability from the technology of measurements). Supervised learning technique based on the Wasserstein metric that is used to estimate an optimal re-weighting of class proportions in a mixture model. Details are presented in Freulon P, Bigot J and Hejblum BP (2021) <arXiv:2006.09003>.
- URL:
-
Contact:
Boris Hejblum
8 New results
8.1 High-dimensional statistical learning
8.1.1 Valid inference in high-dimension
Statistical inference in high dimension remains challenging. In particular, the sound analysis of transcriptomic data requires valid statistical testing procedures with well calibrated control of of the type I error, which we have demonstrated for our developed methods 46. In addition, the analysis of high-dimensional longitudinal and time-to-event data bears its own layer of complexity, and we have extended Random Forests to better accomodate such cases which are common in clinical trials 45, 44, 58.
So-called “double dipping”, i.e. using the same data twice, first to identify clusters and second to identify statistically significant variables diffentiating those clusters, leads to poor statistical performances overall without carefully adjusting for this double step – yet it is widely used in transcriptomic data analysis. We have proposed new tests, both parametric and non-parametric, for such post-clustering inference that all enjoy statistical guarantees in this post-clustering inference setting 37.
A surrogate marker is a marker that can be measured earlier and/or more easily than the original clinical outcome, while retaining the ability to reliably assess the impact of a treatment. Those bear a particular interest in interventional studies (eg vaccine trials) where multiple omics data are measured a few hours or days after the intervention as it could significantly accelerate future studies. We have made new developments towards establishing a method for investigating and validating high-dimensional markers as surrogate 12.
8.1.2 Optimal transport for cell-type proportions inference
Variations of cellular population abundance is of critical importance in following the immune state of vaccine trial participants. It is usually measured from flow-cytometry or mass-cytometry measurements, high-throughput single-cell technologies. Raw output data are manually processed through specific sequences of 2-dimensional projections form measured cellular markers, an expensive, subjective and time-consuming approach. We participated in new developments of optimal transport to infer mixture proportions robust to domain changes 23. Those developments lead to a better characterization and new results regarding budgeted computational efficiency of algorithms estimating regularized Wasserstein distances 40. These algorithms are paramount for the practical application of these statistical tools, in particular to cytometry data.
8.1.3 Machine learning applied to EHR
We have leveraged both public data and EHR data from the Bordeaux Hospital wharehouse in order to better predict the future number of COVID-19 hospitalized patients. We compared several machine learning algorithms including Random Forests as well as penalized likelood, the latter giving the best performing predictions 22.
8.1.4 Microbiota data
Recently, the team has been interested in microbiota data analysis. An article summarizing the current state of knowledge about the association between respiratory microbiota and chronic respiratory diseases has just been published 15.
8.2 Mechanistic learning
8.2.1 Parameters estimation in mechanistic models
The stakes of estimating parameters in non-linear mixed effects models are high as it is a critical step in understanding the underlying processes that generate the data. Accurately estimating the parameters allows for making valid inferences and predictions about the system being studied. Inaccurate parameter estimates can lead to incorrect conclusions and biased results. In particular, existing approach assume that parameters are constant in time (if non dependent on an observed time-varying covariate). This assumption could be relaxed by using approaches based on Kalman filters in which parameters can also have a dynamics. Collin et al. 20 introduce a population-based Karlman filter estimation method. Collin et al. 19 used it to address the problem of estimating non-pharmaceutical intervention effects on COVID19 epidemics.
Another major pitfall of classic methods such as maximum likelihood is their inability to account for model error during parameter inference. This could have a dramatic effect on estimation accuracy. To this end, Clairon et al. 42 proposed an estimation method based on optimal control theory to acknowledge model error presence at the subject level in non-linear mixed effect models.
8.2.2 Model building in non linear mixed effect models
Construction of a complex (nonlinear) mixed-effects model is a challenging process which requires confirmed expertise, advanced statistical methods, and the use of sophisticated software tools, but, above all, time and patience. Indeed, the success of correctly identifying all the components of the model is far from straightforward: it is a question of finding the best structural model, determining the type of relationship between covariates and individual parameters, detecting possible correlations between random effects, or also modeling residual errors. Our goal is to accelerate and optimize this process of model building. In collaboration with L. Lavielle team XPOP, Prague et al. (2022) 31 presents the Stochastic Approximation for Model Building Algorithm (SAMBA) procedure, a method for building nonlinear mixed-effects models that reduces the computational time required for selecting the covariate model. The study compares the performance of SAMBA to existing methods, Stepwise Covariate Modeling (SCM) and COnditional Sampling use for Stepwise Approach (COSSAC), and finds that SAMBA performs similarly while reducing computation time. The study also shows that SAMBA can be used to automatically select correlation and error models in nonlinear mixed-effects models. This method could potentially be applied in drug discovery, development and therapeutics by quickly finding good models that fit the data and are parsimonious in terms of parameters and covariates. Next step could be to extend the approach in high-dimension settings.
8.2.3 Use of mechanistic models to define correlates of protection
The definition of correlates of protection is a difficult task, closely related to causality. Mechanistic model adresses the causality aspects in dynamical data. Thus in Alexandre et al. (2022) 13, we propose a mechanistic model-based approach for identifying correlates of protection for the development of next-generation SARS-CoV-2 vaccines. The approach involves mathematical modeling of viral dynamics and data mining of immunological markers. The model is applied to three studies in non-human primates evaluating different vaccine platforms. The results show that two main mechanisms for protection with vaccine are a decrease in the rate of cell infection and an increase in clearance of infected cells. We show that Inhibition of RBD binding to ACE2 is a robust mechanistic correlate of protection across the three vaccine platforms.Next step could be to extend the approach to find a threshold for protection against transmission.
8.3 Translational vaccinology
A large randomized multi-arm Ebola vaccine trial (Prevac trial, evaluating three different Ebola vaccine strategies 27) has had its primarily results available and a manuscript has been published in the New England Journal of Medicine (December 2022).
The ANRS VRI01 phase I/II vaccine trial, for which we had developed an innovative early phase design [RDL+14] has been completed and final results published 32.
Furthermore, the ANRS VRI06 first-in-human phase I trial of a novel vaccine concept targeting dendritic cells (here as HIV vaccine) has started in 2021, and we have prepared the trial design of next generation Coronavirus vaccines based on that novel concept. W26 results of the Solo groups of the ANRS VRI06 vaccine trial have become available in 2022 and submitted to CROI 2023.
An important milestone is also the set-up and conduct of the national CovPopArt Covid Vaccine Cohort (co-PI: LW): a French cohort for assessing COVID-19 vaccine responses in specific populations, with results becoming available in 2022 29.
Covid treatment trials: The researchers of the axis have been heavily mobilized by the Coverage France clinical trial. Coverage France is a national multi-arm multi-stage (MAMS) adaptive trial platform for early treatments in Covid-infected outpatients in France that has been set-up in spring 2020 and obtained the “national priority” label (Capnet) 66, 56. The trial has been adapted several times since its start, including evolution of the design and the evaluation of new treatment strategies, and is stopped since December 2021. A manuscript with the results of one of the tested treatment strategies (ciclesonide) has been published in 2022 (co-first author 21) and another publication is under preparation.
The PhD project of Cyrille Kone (co-directed with Emilie Kaufmann, Inria Scool) on the development of novel early phase vaccine trial designs based on bandit algorithms started in Nov 2022.
We have set up a data warehouse system based on the Labkey solution where all raw data are organized and that includes meta-data on the design of the clinical trials and is used in international collaborations of facilitate data sharing and exploration (EHVA, EBOVAC, IP-Cure-B and CARE consortia).
Participants: Marta Avalos, Quentin Clairon, Robin Genuer, Boris Hejblum, Edouard Lhomme, Mélanie Prague, Laura Richert, Rodolphe Thiébaut, Linda Wittkop.
9 Partnerships and cooperations
9.1 International initiatives
9.1.1 Associate Teams in the framework of an Inria International Lab or in the framework of an Inria International Program
DESTRIER
-
Title:
DEfining Surrogacy of early Transcriptomics foR vaccInE Response
-
Duration:
2022 ->
-
Coordinator:
Denis Agniel (dagniel@rand.org)
-
Partners:
- RAND Corporation (États-Unis)
-
Inria contact:
Boris Hejblum
-
Summary:
This project seeks to develop statistical methods to evaluate to which extent can transcriptomics be used to capture vaccine effects. Gene expression is central to protein production and largely determines cellular function: it is thus a promising biomarker for quickly measuring effects of vaccines. Validated transcriptomic signatures could thus be developed to dramatically speed up vaccine trials for emerging infectious diseases like Ebola or COVID-19. Such a technology could also be used for identifying good vaccine responders in health care workers that can be deployed in case of an epidemic emergency, or identify poor responders among vaccine recipient that would benefit from an additional booster dose. In this project, we set to develop novel statistical methods for assessing the surrogacy potential of transcriptomic data in vaccine research. We will first develop methods to quantify how much of the vaccine effect is mediated by gene expression, establishing if gene expression is suitable for capturing the vaccine's effect. We will develop model-free approaches to estimating this quantity which will remove many of the modeling assumptions typically used in high-dimensional mediation analysis. Second, we will develop methods to construct an optimal gene expression signature for capturing the vaccine effect, and we will develop methods to operationalize its use in future studies, establishing how to build and use such a transcriptomic signature. These methods will similarly take advantage of modern machine learning approaches and doubly robust estimation to provide model-free estimators of key quantities. We will use these methods to study high-impact clinical trials from the Vaccine Research Institute in the context of Ebola and COVID-19.
DYNAMHIC
-
Title:
DYNAMical modeling of HIV Cures
-
Duration:
2019 ->
-
Coordinator:
Alison HILL (alhill@fas.harvard.edu)
-
Partners:
- Harvard University Cambridge (États-Unis)
-
Inria contact:
Melanie Prague
-
Summary:
The aim of the DYNAMHIC Associate Team is to bring together a mathematical biology team at Harvard and the Inria team SISTM of applied statisticians at Bordeaux Sud-ouest. This collaboration will allow the analysis of unique pre-clinical non human primates data of HIV cure interventions. In particular, we will focus on immunotherapy and therapeutic vaccine, which are very promising in term of efficacy and are at the leading edge of pre-clinical research in the area. The novelty of the approach is to propose an integrative project studying complex biological processes with novel mathematical statistical models, which has the potential to yield predictive computational tools to assist in the design of both therapeutic products and clinical trials for HIV cure
Finally, the associate team is the opportunity to provide the research group with an official administrative framework. And, to continue to develop a promising research topic connected but different from those funded up to now.
9.1.2 Participation in other International Programs
NIPAH (Chine) – Scientific cooperation program France/Chine. (2019-2022). M. Prague is workpackage co-PI - Sino-French Agreement Aviesan. Sep 2018 – Aug 2023, 150,000 euros. To raise the challenge caused by Nipah virus we propose to develop a program that shall led to a better understanding of the epidemiology of the virus as well as the associated physiopathology. To develop new tools in the field of diagnosis, treatment and prevention of the infection. This grant aims at funding a 2 years of postdoc, travel and equipment expenses.
9.2 International research visitors
9.2.1 Visits of international scientists
Other international visits to the team
Daniel Dori
-
Status
PhD student
-
Institution of origin:
Université Joseph Ki-Zerbo
-
Country:
Burkina Faso
-
Dates:
from Jun 1st 2022 until Jun 30th 2022
- Context of the visit:
-
Mobility program/type of mobility:
internship
Jane Heffernan
-
Status
Professor
-
Institution of origin:
York University of Toronto
-
Country:
Canada
-
Dates:
from May 1st 2022 until May 30th 2022
-
Context of the visit:
Mathematical modelling
-
Mobility program/type of mobility:
Invited Professor
Isabela Ornelas
-
Status
Epidemiologist
-
Institution of origin:
Brasilian Ministry of Health
-
Country:
Brasil
-
Dates:
from Oct 17th 2022 until Dec 15th 2022
- Context of the visit:
-
Mobility program/type of mobility:
Research visit
Baptiste Elie
-
Status
PhD student with Samuel Alizon (Collège de France)
-
Institution of origin:
Ecole Normale Supérieure Paris-Saclay
-
Country:
France
-
Dates:
from Jun 1st 2022 until Jun 30th 2022
-
Context of the visit:
Modeling HPV dynamics
-
Mobility program/type of mobility:
Travel grant / scientific exchange ANRS MIE
9.3 European initiatives
9.3.1 H2020 projects
-
IP-CURE-B: Immune profiling to guide host-directed interventions to cure HBV infections. Co-ordinated by Inserm (France, the project includes a total of 13 Beneficiaries: Centre Hospitalier Universitaire Vaudois (Switzerland), Karolinska Institutet (Sweden), Institut Pasteur (France), Universita degli studi di Parma (Italy), Fondazione IRCCS CA’ Granda – Ospedale maggiore policlinico (Italy), Universitaet- sklinikum Freiburg (Germany), Ethniko Kai Kapodistriako Panepistimio Athi-non (Greece), Fundacio Hospital Universitari vall d’Hebron (Spain), Gilead Sciences Inc. (USA), Spring Bank Pharmaceuticals, Inc (USA), European Liver Patients Association (Belgium), Inserm Transfert SA (France). L Wittkop. Duration: 60 months 01/01/20-31/12/24. 409,632 Euros. -
EHVA (European HIV Vaccine Alliance): European HIV Vaccine Alliance: a EU platform for the discovery and evaluation of novel prophylactic and therapeutic vaccine candidates. Coordinator: Inserm/University of Lausanne. Other partners: EHVA consortium gathers 41 partners. R. Thiébaut. Duration: 60 months. 01/01/2016 - 30/06/23 – 208 686 euros. -
CoVICIS (EU-Africa Concerted Action on SAR-CoV-2 Virus Variant and Immunological Surveillance): The CoVICIS program is proposing a global approach with a powerful state-of-the-art virologic and immunologic platforms coupled with large genomic surveillance studies and diverse cohorts in EU and SSA CoVICIS aims to contribute to the early identification of emerging VOC and address key unanswered questions regarding: i) the susceptibility to infection with VOC after a prior infection in the setting of a long-COVID or after vaccination with different vaccines, ii) the risk posed by VOC in immunocompromised patients, and iii) the modalities of infection and immune responses in children. CoVICIS is coordinated by the CHUV (Switzerland). SISTM is involved in WP7 Data Science and Analysis which aims to utilize cutting edge computational and statistical analysis method to obtain comprehensive assessment of immunogenicity and immune correlates of protection. 11/2021-10/2024. Coordinated by the CHUV, CoVICIS counts 14 partners amongst which we can find Inserm, UNIMI, UNIGE, and 4 South-African partners. Total budget: 10M€, SISTM budget: 110k€.
9.3.2 Other European programs/initiatives
- EBOVAC1: Development of a Prophylactic Ebola Vaccine Using an Heterologous Prime-Boost Regimen. Coordinated by London School of Hygiene & Tropical Medicine (United Kingdom). Other beneficiaries: Janssen a Pharmaceutical Companies of Johnson and Johnson, The Chancellor, Masters and Scholars of the University of Oxford (United Kingdom), Inserm (France), University of Sierra Leone (Sierra Leone), R. Thiébaut. Duration: 84 months. 01/12/2014 - 31/05/2023. 552,050 Euros.
- EBOVAC3: Bringing a prophylactic Ebola vaccine to licensure. Coordinated by the London School of Hygiene & Tropical Medicine (United Kingdom). Other beneficiaries: Janssen a Pharmaceutical Companies of Johnson and Johnson, Inserm (France), The University of Antwerpen (Belgium), University of Sierra Leone (Sierra Leone), R. Thiébaut. Duration: 60 months. 01 /06 /2018 - 30 /06 /2023. 351,274 Euros.
- PREVAC-UP: The Partnership for Research on Ebola VACcinations-extended follow-UP and clinical research capacity build-UP. SISTM is also involved in PREVAC-UP, an EDCTP2 project in direct link with the research carried out on the Ebola vaccines. Coordinated by Inserm (France). Other beneficia- ries: CNFRSR (Guinea), CERFIG (Guinea), LSHTM (UK), COMAHS (Sierra-Leone), NIAID (USA), NPHIL, (Liberia), USTTB (Mali), Centre pour le Développement des Vaccins (Mali), Inserm Transfert SA (France), R. Thiébaut. Duration: 60 months. 01 /01 /2019 - 31 /12 /2023. 328,000 Euros.
- CARE: Corona Accelerated R&D in Europe is an IMI2 funded project coordinated by Inserm which gathers 36 globally renowned academic institutions, pharmaceutical companies and non-profit research organisations which have committed to rapidly and efficiently address the COVID-19 emergent heath threat. This major initiative aims at addressing two key objectives: the development of therapeutics to provide an emergency response towards the current COVID-19 pandemic and the development of therapeutics to address the current and/or future coronavirus outbreaks. To address both goals, the CARE consortium has carefully designed a comprehensive research and development (R&D) program around thoughtfully designed Target Product Profiles (TPP) of the urgently needed antiCOVID-19 drugs. This includes small and large molecule discovery and Phase 1 and 2 clinical trials centred around three main pillars: drug repositioning, small-molecule drug discovery, and virus neutralising antibody discovery. These pillars reflect a bifocal strategy where efforts are geared towards (a) a rapid response against current COVID-19 pandemic and (b) a longer-term preparedness strategy against future coronavirus outbreaks. This will maximize the screening landscape of relevant therapeutic avenues and ensure effective therapeutics can be rapidly identified, pre-clinically tested and optimised for clinical-grade manufacturing and clinical testing. In this project, SISTM and EUCLID are working closely together with the support of the CREDIM in the WP5, W7 and WP8 with the respective objectives of providing statistical analysis and data modelling of the immune assays carried out in the project, bring some expert support to the clinical work and develop a LabKey-based platform for the integration and management of the data. Duration: 60 months. 01/04/2020 - 30/03/2025. 1,256,003 Euros.
- ASCENT: Acceleration of Novel Coronavirus Serological Test Development and Seroprevalence Study: An African-European Initiative. ASCENT is an EDCTP2 projects involving 7 partners (Inserm, CHUV, EuroVacc, Utrecht University, Centre Muraz, SAMRC and CERFIG) from 6 different countries in Africa and Europe which will aim at assessing the real prevalence of the infection, the projection of the immunity acquired by the populations, and the evaluation of measures aimed to break the transmission in Africa. To do so ASCENT will implement in Burkina Faso, South Africa and Guinea, a novel robust and reproducible luminex-based serological diagnostic test with high throughput, sensitivity, specificity and rapid turn- around time. In this project, SISTM will be involved in statistical analysis of the tests data and will lead the WP3 which aims at modelling the epidemics. Duration: 24 months. 01/05/2020 - 30/04/2022. 37,500 Euros.
- CoVICIS (EU-Africa Concerted Action on SAR-CoV-2 Virus Variant and Imunological Surveillance): The CoVICIS program is proposing a global approachwith a powerful state-of-the-art virologic and immunologic platforms coupled withlarge genomic surveillance studies and diverse cohorts in EU and SSA CoVICIS aims to contribute to the early identification of emerging VOC and address key unanswered questions regarding: i) the susceptibility to infection with VOC after a prior infection in the setting of a long-COVID or after vaccination with different vaccines, ii) the riskposed by VOC in immunocompromised patients, and iii) the modalities of infectionand immune responses in children. CoVICIS is coordinated by the CHUV (Switzerland). SISTM is involved in WP7 Data Science and Analysis which aims to utilize cutting edge computational and statistical analysis method to obtain comprehensive assessment of immunogenicity and immune correlates of protection. Start 11/2021 End 10/2024 Main partners: Coordinated by the CHUV, CoVICIS counts 14 partners amongst which we can find Inserm, UNIMI, UNIGE, and 4 South-African partners Total budget: 10MC, SISTM budget: 110kC.
9.4 National initiatives
-
Labex Vaccine Research Institute (
VRI): Funded by the PIA under Laboratory of excellence initiative, VRI conducts research to accelerate the development of effective vaccines against HIV/AIDS and (re)-emerging infectious diseases. The SISTM team is leading the Data science division of the VRI. To this purpose, SISTM has established strong collaboration with immunologists. SISTM carries out biostatistical analysis of the data produced by the different other VRI teams together with a modelling approach of the immune response to the vaccines or other interventions. 2012-2024, Main partners: the VRI was established by the French National Agency for Research on AIDS and viral hepatitis (ANRS – France Recherche Nord & Sud Sida-HIV Hépatites) and the University of Paris-Est Créteil (UPEC). The other partners of the VRI are CEA, Inserm, Pasteur Institute, the University of Bordeaux, the Baylor Institute for immunology research and the University of Strasbourg. Total budget: 75M€, SISTM budget: 1.85M€ (about 170k€ a year since 2012). -
RHU
SHIVA: Since November 2019, the RHU SHIVA aims to better understand the determinants and consequences of cerebral small vessel disease through innovative imaging, molecular and analytical approaches, to develop new personalized diagnostic and preventive strategies and to accelerate the discovery of novel therapeutic targets. RT as workpackage leader is collaborating in the SHIVA RHU to work on the integration and systems biology of small vessel disease biomarkers. 12/2019-11/2024. Main partners: University of Bordeaux, Inserm, CNRS, CHU Bordeaux, CHNO AP-HP, Fealinx, Qynapse, ImaginEyes. Total budget: 8.2 M€, SISTM budget: 190k€. -
Ecole Universitaire de Recherche
“Digital Public Health” Funded under the PIA3 The Digital Public Health Graduate Program provides an interdisciplinary and international training from Master to Doctorate in epidemiology, biostatistics, computing and social sciences to explore the impact of digital public health on society. The whole program is directed by Rodolphe Thiébaut. The whole SISTM team is implicated in these activities. 2018-2028. Main partners: University of Bordeaux, Inserm, Inria, Sciences Po Bordeaux and University Bordeaux Montaigne. Total budget: 4.52 M€, SISTM budget: The budget is mostly dedicated to grants to students, running costs and indemnification of teachers. -
Project Emergen: Consortium for Surveillance and Research on EMERgent Pathogens via Microbial GENomics. EMERGEN, coordinated by Sante publique France and ANRS-Emerging Infectious Diseases, aims to deploy a genomic surveillance system for SARS-CoV-2 infections throughout France. Its main objective is to to follow the genetic evolution of the SARS-CoV-2 virus in order to detect the emergence and the spatio-temporal distribution of variants, i.e. viruses with mutations likely to have functional consequences, such as infectivity, contagiousness, virulence or immune escape. In this context, the role of SISTM will be to contribute to modelling the impact on epidemic dynamics of SARS-CoV-2 variants based on the estimation of their characteristics. 01/2022-01/2024. Main partners: Santé Publique France, Inserm/ANRS, APHP, HCL, Pasteur Institute, Anses, IFB, CNRGH/CEA, Réseau Sentinelles. Total budget: 10M€, SISTM budget: 56k€.
9.4.1 Various Partnership
Mélanie Prague: Chaire Digital Innovation and Health Data Science program of the Center for Applied Mathematics CMAP at the Ecole Polytechnique
The project team members are involved in:
- F-CRIN (French clinical research infrastructure network), initiated in 2012 by ANR under "Programme des Investissements d'avenir". (L Richert).
- Contrat Initiation ANRS MoDeL-CI: Modeling the HIV epidemic in Ivory Coast (Principal PI Eric Ouattara Inserm U1219 in collaboration with University College London, Mélanie Prague is listed as a collaborator).
- TARPON (Traitement Automatique des Résumés de Passages aux urgences pour un Observatoire National), laureate project from the 2nd Health Data Hub calls for projects, great challenge "Improving medical diagnostics through Artificial Intelligence" and Bpifrance. (Principal PI E. Lagarde Inserm U1219 in collaboration with University Hospital of Bordeaux. Marta Avalos is listed as a collaborator).
- TACAUD (Traitement Automatique des données du Centre de régulation des Appels d’Urgence Départemental), (Principal PI E. Lagarde Inserm U1219 in collaboration with University Hospital of Bordeaux. Marta Avalos is listed as a collaborator).
- CESIR IV (Combination of Studies on Health and Road Safety - 4th project) funded by ONISR DSR. (Principal PI E. Lagarde Inserm U1219. Marta Avalos is listed as a collaborator).
- Collaboration with Inserm PRC (pôle Recherche clinique).
- Collaboration with Inserm REACTing (REsearch and ACTion targeting emerging infectious diseases) network.
- Collaboration with Inserm RECap (Recherche en Epidémiologie Clinique et en Santé Publique) network.
- STRIVE (Strategies and Treatments for Respiratory and Viral Emergencies Study Payments). International Network for respiratory and viral emergency studies. (Collaborator: Linda Wittkop).
Participants: Marta Avalos, Quentin Clairon, Robin Genuer, Boris Hejblum, Edouard Lhomme, Mélanie Prague, Laura Richert, Rodolphe Thiébaut, Linda Wittkop.
10 Dissemination
10.1 Promoting scientific activities
10.1.1 Scientific events: organisation
General chair, scientific chair
Mélanie Prague organized the ANRS MIE days Modelling of infectious diseases – Bordeaux, Nov 20-21 2022 Acmodelling-22.
Boris Hejblum organizes the Biostatistics Seminar Series at the Bordeaux Population Health Inserm Research Center.
Member of the organizing committees
Sandrine Darmigny, Mélanie Prague, Quentin Clairon and Marie Alexandre organized the logistic for the ANRS MIE days Modelling of infectious diseases – Bordeaux, Nov 20-21 2022 (80 people).
Mélanie Prague organized a full-day in person (no hybrid option) meeting for the working group "within-host" of the AC modelling of ANRS MIE. In Paris Bichat university in collaboration with Jérémie Guedj (60 persons). Alan Perelson was keynote speaker.
10.1.2 Scientific events: selection
Chair of conference program committees
Rodolphe Thiébaut: chair of the scientific commitee of the 9th International Meeting on Statistical Methods in Biopharmacy, Paris 19-21 Sep 2022.
Rodolphe Thiébaut: chair of the scientific commitee of the of the IWHOD International Workshop on HIV Observational Databases in 2020-2022
Member of the conference program committees
Marta Avalos was a member of the Program Committee of
- the ACM Conference on Health, Inference, and Learning - CHIL 2022 (from 2020)
- the Machine Learning for Health – ML4H Workshop at NeurIPS 2022 (from 2019)
- Conférence sur l'apprentissage automatique - CAp 2022 (from 2018), and
- Dataquitaine 2022 IA, Recherche Opérationnelle & Data Science.
Rodolphe Thiébaut was a member of the scientific committee of the
- national conference on clinical research (EPICLIN) from 2017 and Laura Richert from 2020.
- IWHOD International Workshop on HIV Observational Databases since 2013 IWHOD.
Laura Richert is a member of the scientific committee of the national conference on clinical research (EPICLIN) from 2020.
10.1.3 Journal
Member of the editorial boards
Mélanie Prague is associate editor of "International journal of Biostatistics" (since 2018).
Rodolpe Thiébaut is section editor of IMIA Yearbook in Medical Informatics (since 2017).
Reviewer - reviewing activities
- AIDS, Journal of Immunology, Nature communications (Rodolphe Thiébaut)
- Annals of Applied Statistics (Boris Hejblum)
- Antiviral Therapy (Linda Wittkop)
- Biometrics (Mélanie Prague, Boris Hejblum, Rodolphe Thiébaut)
- Bayesian Analysis (Boris Hejblum)
- Bioinformatics (Boris Hejblum)
- Computational Statistics and Data Analysis (Boris Hejblum)
- Eletronic Journal of Statistics (Robin Genuer)
- Gut (Linda Wittkop)
- IMIA Yearb Med Inform (Marta Avalos)
- Journal of Computation Statistics and Data Analysis (Boris Hejblum)
- Journal of Hepatology (Linda Wittkop)
- Journal of International Medical Research (Marta Avalos)
- PLOS Computational Biology (Boris Hejblum, Mélanie Prague)
- STAT (Boris Hejblum)
- Statistics in Medicine (Mélanie Prague, Rodolphe Thiébaut)
- Statistical science (Mélanie Prague)
- Trials (Laura Richert, Rodolphe Thiébaut)
10.1.4 Invited talks
Mélanie Prague:
- Prague M., Alexandre M., Marlin R, Le grand R, Levy Y, Thiébaut R. Elicitation of SARS-CoV-2 mechanistic correlates of protection using mechanistic models. The Canadian Applied and Industrial Mathematics Society 13-16 June Hybrid.
- M. Prague, M. Alexandre, R. Rhiébaut SARS-CoV-2 mechanistic correlates of protection Rennes INSA, June
Linda Wittkop:
- Vaccelerate seminar/webinar : Humoral immune response after COVID-19 vaccination in specific populations – ANRS0001S COV POPART cohort study, 12 April 2022
- ELPA seminar (1st leymen event): Importance of clinical trials and their meth-odology for the demonstration of a drug activity, 23 November 2022
- BPH seminar: COVID-19 Vaccination in specific populations: results from the ANRS0001S COV-POPART study, 18 November 2022
- EUCLID seminar : the ANRS0001S COV-POPART study: Challenges of setting up a Covid-19 vaccine cohort for special populations in times of a health crisis, 24 November 2022.
- COVIREIVAC seminar: Presentation of the ANRS0001S COV-POPART, Annecy, France, 7 March 2022
- COVIREIVAC webinar for participants : First results from the ANRS0001S COV-POPART study, 8 October 2022
Rodolphe Thiébaut:
- Joint Annual Meetings of the French Society for Immunology and the French Cytometry Association November 22-24, 2022, Nice, France, Analysis of gene expression data applied to SARS-COV2 infected patients
- IVI’s 21st International Vaccinology Course September 26-30, 2022, Stockholm, Sweden, Mathematical modelling of infectious diseases
- Paris Symposium on viral dynamics September 16, 2022, Paris, Modelling B cell response to vaccine. Exemple of Ad26/MVA against Ebola virus disease
- Intelligence artificielle et santé : approches interdisciplinaires – Jun 29th -Jul 1st 2022, Nantes, Analyzing transcriptomics data for understanding and predicting vaccine response in clinical trials
10.1.5 Scientific expertise
- Rodolphe Thiébaut is a member of the Pasteur Institute evaluation committee, a member of the independent committee of international trials ODYSSEY, SMILE, BREATHER, 3D. He is the coordinator of the evaluation committee for the Italian « “Extended Partnerships with universities, research centres and companies to fund basic research projects” - within the framework of the National Recovery and Resilience Plan (160 M€ over 3 years).
- Edouard Lhomme is an expert for the PHRC (Programme hospitalier de recherche Clinique).
- Laura Richert is an expert for the PHRC (Programme hospitalier de recherche Clinique) and a member of the CNU 46.04 (Biostatistiques, informatique médicale et technologies de communication).
- Linda Wittkop is a member of the external ethics and scientific advisory board of the EU-funded project VACCELERATE and a member of the scientific commitee CSS 13 (Recherches cliniques et physiopa-thologiques dans l’infection à VIH) of the ANRS (France Recherche Nord& Sud Sida-HIV Hépatites) and a member of the CESREES (Comité éthique et scientifique pour les Recherches, les Etudes et les Evaluation dans le domaine de la santé).
- Mélanie Prague is an expert for ANRS (France Recherche Nord&Sud Sida-HIV Hépatites) in the CSS13 (Recherches cliniques et physiopathologiques dans l’infection à VIH).
10.1.6 Research administration
Rodolphe Thiébaut is a member of the Scientific Council of Inserm (since 2017)
Boris Hejblum is a member of the chairing committee of the Société Française de Biométrie, the French Chapter of the International Biometric Society and a board member of the “MAchine Learning et Intelligence Artificielle” (MALIA) group of the French Society of Statistics (SFdS).
Hélène Savel is a board member of the “Biopharmacie et Santé” group of SFdS.
Mélanie Prague is in the Bureau of the “Action Coordonnée Modélisation des maladies infectieuses" (ANRS MIE) and she is co-president of the communication group of SFdS, in charge of redefining de condition of sponsoring SfdS by enterprises. She is a member of the computational means users committee and took part of the jury Inria Saclay 2022.
Edouard Lhomme is co-president of the joint action on respiratory viruses of ANRS-MIE.
Laura Richert is coordinator of the Clinical epidemiology module of the Clinical Investigations Center (CIC1401 Bordeaux).
Linda Wittkop is director of the mixed Inserm/Bordeaux university unit UMS 54 Methods and Applied research of Trials and coordinator of the axis “Infectious diseases and Inflammation” of the CIC1401-E.
10.2 Teaching - Supervision - Juries
10.2.1 Teaching
Each staff member is involved in teaching with approximatively MA 200 h/year, RG 200 h/year, BH 70 h/year, EL 80 h/year, MP 70 h/year, LR 80 h/year, RT 130 h/year, and LW 110 h/year. These activities splits as follow.
-
In class teaching
- Rodolphe Thiébaut is head of the Digital Public Health graduate program, University of Bordeaux. Robin Genuer is head of the M2 Biostatistique, Master of Public Health, University of Bordeaux.
- Master: All the permanent members and several PhD students teaches in the Master of Public Health (M1 Santé publique, M2 Biostatistique and/or M2 Epidemiology) and the Digital Public Health graduate program, University of Bordeaux.
- Master: Marta Avalos teaches in the Master of Applied Mathematics and Statistics (1st and/or 2nd year), University of Bordeaux.
- Bachelor: Laura Richert, Linda Wittkop and Edouard Lhomme teach in PASS and DFASM1-3 (Diplôme de Formation Approfondie en Sciences Médicales) for Medical degree at Univ. Bordeaux.
- Master: Laura Richert and Edouard Lhomme teach in the Master of Vaccinology from basic immunology to social sciences of health (University Paris-Est Créteil, UPEC).
- Teaching unit coordination: Laura Richert, Linda Wittkop, Rodolphe Thiébaut, Robin Genuer, Boris Hejblum and Marta Avalos coordinate several teaching units of Master in Public Health (Biostatistics, Epidemiology, Public Health).
- Laura Richert coordinates the teaching unit "critical article reading" (across 4 years of medical school), University of Bordeaux; Edouard Lhomme coordinates the teaching “Evaluation of health innovation (M2 Health innovations); Linda Wittkop coordinates the teaching unit “Public Health and Statistics in Medicine” of the first year of Medical School, University of Bordeaux; Marta Avalos co-coordinates the teaching unit "Statistical analysis of high-dimensional data" (M2 Statistical and stochastic modeling MSS and M2 in Statistical and Computer Engineering CMI ISI of the UFMI of the University of Bordeaux).
- Summer school: Mélanie Prague teaches in the summer school, University of Bordeaux.
- Boris Hejblum teaches a 3-day graduate course “Bayesian analysis for biomedical research” at the University of Copenhaguen.
- Mélanie Prague teaches "Missing Data" at ENSAI Master Level and Mixed effects models for populational approaches at Ecole polytechnique M2 data science.
- ISPED Summer school: Introduction to R (Mélanie Prague), Advanced R (R Genuer & B Hejblum).
- Training courses in clinical trials in Guinea (Edouard Lhomme).
-
E-learning
- Marta Avalos is head of the e-learning program of the Master of Public Health, 1st year, University of Bordeaux.
- Master: Marta Avalos teaches in the e-learning program of the Master of Public Health (1st and 2nd year).
- ODL University Course: Robin Genuer is head of the Diplôme universitaire "Méthodes statistiques en santé". Mélanie Prague teaches in the Diplôme universitaire "Méthodes statistiques de régression en épidémiologie".
- ODL University Course: Edouard Lhomme co-coordinates and teaches in the Diplôme universitaire "Recherche Clinique".
- ODL University project: Robin Genuer participated to the IdEx Bordeaux University "Défi numérique" project BeginR.
10.2.2 Supervision
PhD defense: Anthony Devaux defended his thesis in biostatistics, untitled “Dynamic modelling and prediction of health events from multivariate longitudinal data” and co-directed by Cécile Proust-Lima and Robin Genuer, on the 29/11/2022, Bordeaux University.
PhD defense: Marie Alexandre “Mechanistic modeling of vaccine response”, directed by Mélanie Prague and Rodolphe Thiébaut, 10/05/2022, Bordeaux University.
PhD in progress: Houreratou Barry “Response to Ebola vaccine and factors associated with the variablity of vaccine immune response in African countries” -directed by Rodolphe Thiébaut, from Oct 2019
PhD in progress: Iris Ganser, Evaluation of event-based internet biosurveillance for multi-regional detection of seasonal influenza onset, co-directed by David Buckeridge (McGill University) and Rodolphe Thiébaut, from Oct 2020.
PhD Ipsen, CIFRE in progress: Hélène Savel "Statistical analysis of OMICS data for the treatment response prediction in early clinical development in a context of the generation of virtual patients to run In Silico Clinical Trials", directed by Laura Richert, from Oct 2020.
PhD in progress: Benjamin Hivert "Hierarchical modeling for integrative analysis of high-dimensional, high-throughput, multi-modal cell and molecular data for immunology research", co-directed by Boris Hejblum and Rodolphe Thiébaut, from Sept 2020.
PhD in progress: Thomas Ferte "Contribution of health data warehouses for clinical research and epidemiological surveillance in the context of Covid-19", co-directed by Rodolphe Thiébaut and Vianney Jouhet, from Sept 2021.
PhD in progress: Kalidou Ba “Reservoir computing for cellular composition prediction from longitudinal transcriptomics data in vaccine trials”, co-directed by Rodolphe Thiébaut and Xavier Hinaut, from Nov 2022.
PhD in progress: Cyrille Kone “Bandit algorithms for early phase clinical trials in vaccinology”, co-directed by Emilie Kaufmann (Inria Scool) and Laura Richert from November 2022.
Marta Avalos is the academic tutor of PhD student S. Zaïd, Ecole doctorale Sociétés, Politique, Santé Publique de l'Université de Bordeaux- EDSP2.
Master students:
- Mélanie Prague supervised the internship of Florian Robert (M2 Sorbonne) and Marie Poupelin (M1 ISPED, Université de Bordeaux)
- Marta Avalos supervised the internship of Vishwa Elankumaran (M2 Université de Lyon, co-supervised with Laurence Delhaes, Inserm U1045) and Samia Mehouachi (M1 Santé publique e-learning, ISPED, Université de Bordeaux) and was the academic tutor of the internship of Grégory Arrotis (M2 MSS, Université de Bordeaux) at KPC Bordeaux.
- Laura Richert and Edouard Lhomme supervised the internship of Simon Valayer (M2) and Bhushan Rugbursing (DFGSM-3).
- Quentin Clairon supervised the internship of Justine Remiat (M1).
- Linda Wittkop supervised the internship of Raphaël Garcia (M1).
10.2.3 Juries
Jury HDR:
- Yusuke Shimakawa, Institut Pasteur, L Wittkop
- Amadine Crombé, Applied Mathematics, the 13/12/2022, Bordeaux University, Robin Genuer
Jury PhD defense:
- Charlotte Lanièce Delauny, McGill, L Wittkop
- Guillaume Lingas Univ. Paris Diderot, M Prague
Jury PhD Follow-up:
- Houreratou Barry, Université de Bordeaux – L Wittkop, L Richert
- Manel Rakez, Université de Bordeaux – Boris Hejblum
- Baptiste Elie, Université de Bordeaux – Mélanie Prague
Jury MD defense: Idoia Necrete, Université de Bordeaux - L Wittkop.
Jury Master defense: all the members participated to the juries of internship defense of the Master of Public Health (1st and 2nd years, Digital Public Health, Biostatistics, Epidemiology, Public Health e-learning), University of Bordeaux. Marta Avalos participated to the juries of internship defense of the M2 Statistical and stochastic modeling MSS and M2 in Statistical and Computer Engineering CMI ISI of the UFMI of the University of Bordeaux.
Jury recruitment of an assistant professor:
- Rodolphe Thiébaut was the president and Marta Avalos was a member of the jury recruitement of an assistant professor in biostatistics at University of Bordeaux.
- Robin Genuer was a member of the jury recruitement of an assistant professor in statistics and psychology at University of Nantes.
10.3 Popularization
10.3.1 Education
Boris Hejblum animated two “Chiche: Un scientifique, Une classe” at Lycée François Magendie (Bordeaux).
10.3.2 Interventions
Interview with Professor Rodolphe Thiébaut broadcast on France Inter during the Leem press conference: "In 2030, will digital technology have completely revolutionized clinical trials?"
Rodolphe Thiébaut gave a talk in the Bordeaux PharmacoEpi Festival, 10th edition, May 18th to May 20th 2022, "Data science for vaccine development".
Contribution to Dataquitaine 2022 51.
Participants: Marta Avalos, Quentin Clairon, Robin Genuer, Boris Hejblum, Edouard Lhomme, Mélanie Prague, Laura Richert, Rodolphe Thiébaut, Linda Wittkop.
11 Scientific production
11.1 Major publications
- 1 articleVariance component score test for time-course gene set analysis of longitudinal RNA-seq data.Biostatistics1842017, 589-604
- 2 articleSafety and immunogenicity of 2-dose heterologous Ad26.ZEBOV, MVA-BN-Filo Ebola vaccination in healthy and HIV-infected adults: A randomised, placebo-controlled Phase II clinical trial in Africa.PLoS Medicine1810October 2021, e1003813
- 3 articlecytometree: A binary tree algorithm for automatic gating in cytometry analysis.Cytometry Part A93112018, 1132-1140
- 4 bookDynamical Biostatistical Models.Chapman and Hall/CRC2015
- 5 articleModeling CD4 + T cells dynamics in HIV-infected patients receiving repeated cycles of exogenous Interleukin 7.Annals of Applied Statistics2017
- 6 articleA French cohort for assessing COVID-19 vaccine responses in specific populations.Nature Medicine278July 2021, 1319-1321
- 7 articleControlling IL-7 injections in HIV-infected patients.Bulletin of Mathematical Biology2018
- 8 articleSafety and immunogenicity of a two-dose heterologous Ad26.ZEBOV and MVA-BN-Filo Ebola vaccine regimen in adults in Europe (EBOVAC2): a randomised, observer-blind, participant-blind, placebo-controlled, phase 2 trial.The Lancet Infectious DiseasesNovember 2020
- 9 articleDynamic models for estimating the effect of HAART on CD4 in observational studies: Application to the Aquitaine Cohort and the Swiss HIV Cohort Study.Biometrics2017
- 10 articleSystems Vaccinology Identifies an Early Innate Immune Signature as a Correlate of Antibody Responses to the Ebola Vaccine rVSV-ZEBOV.Cell Reports 2092017, 2251 - 2261
- 11 articleAdaptive protocols based on predictions from a mechanistic model of the effect of IL7 on CD4 counts.Statistics in Medicine3822018, 221-235
11.2 Publications of the year
International journals
International peer-reviewed conferences
Conferences without proceedings
Doctoral dissertations and habilitation theses
Reports & preprints
Other scientific publications
11.3 Other
Scientific popularization
11.4 Cited publications
- 52 articleCausality, mediation and time: a dynamic viewpoint.Journal of the Royal Statistical Society: Series A (Statistics in Society)17542012, 831-861
- 53 articleAdaptive Mixture Discriminant Analysis for Supervised Learning with Unobserved Classes.Journal of Classification3112014, 49--84
- 54 unpublishedFréchet random forests for metric space valued regression with non Euclidean predictors.December 2020, working paper or preprint
- 55 articleInhaled ciclesonide for outpatient treatment of COVID-19 in adults at risk of adverse outcomes: a randomised controlled trial (COVERAGE).Clinical Microbiology and InfectionMarch 2022
- 56 articleHome treatment of older people with symptomatic SARS-CoV-2 infection (COVID-19): a structured summary of a study protocol for a multi-arm multi-stage (MAMS) randomized trial to evaluate the efficacy and tolerability of several experimental treatments to reduce the risk of hospitalisation or death in outpatients aged 65 years or older (COVERAGE trial).Trials2112020, 1--3
- 57 articleAutomatic phenotyping of electronical health record: PheVis algorithm.Journal of Biomedical Informatics1172021, 103746
- 58 phdthesisContributions to Random Forests Methods for several Data Analysis Problems.Université de BordeauxJanuary 2021
- 59 articleProbabilistic record linkage of de-identified research datasets with discrepancies using diagnosis codes.Scientific Data 62019, 180298
- 60 articleSystems approaches to biology and disease enable translational systems medicine.Genomics Proteomics Bioinformatics1042012, 181--5
- 61 articleModeling CD4+ T cells dynamics in HIV-infected patients receiving repeated cycles of exogenous Interleukin 7.The Annals of Applied Statistics1132017, 1593--1616
- 62 articleA Benchmark for RNA-seq Deconvolution Analysis under Dynamic Testing Environments.Genome Biology2212021, 102
- 63 articleCoupling a stochastic approximation version of EM with an MCMC procedure.ESAIM: Probability and Statistics82004, 115--131
- 64 bookMixed Effects Models for the Population Approach: Models, Tasks, Methods and Tools.Chapman and Hall/CRC2014
- 65 articleTranslocated Microbiome Composition Determines Immunological Outcome in Treated HIV Infection.Cell184152021, 3899-3914.e16
- 66 articleRandomized trial to evaluate the safety and efficacy of outpatient treatments in individuals with Covid-19 with risk factors. Coverage France trial: a structured summary.Exercer1782021, 451-458
- 67 articleIntroduction to modeling viral infections and immunity.Immunological Reviews28512018, 5-8
- 68 articlePersonalized vaccinology: a review.Vaccine36362018, 5350--5357
- 69 articleDynamic models for estimating the effect of HAART on CD4 in observational studies: Application to the Aquitaine Cohort and the Swiss HIV Cohort Study.Biometrics731March 2017, 294 - 304
- 70 articleNIMROD: A program for inference via a normal approximation of the posterior in models with random effects based on ordinary differential equations.Computer Methods and Programs in Biomedicine11122013, 447--458
- 71 articleLearning immunology from the yellow fever vaccine: innate immunity to systems vaccinology.Nature Reviews Immunology9102009, 741-7
- 72 articleMonolix version 2021R1.Antony, France2022, http://lixoft.com/products/monolix/
- 73 articleSystems immunology: complexity captured.Nature47373452011, 113-4
- 74 articlePheProb: probabilistic phenotyping using diagnosis codes to improve power for genetic association studies.Journal of the American Medical Informatics AssociationMay 2018
- 75 phdthesisRégression pénalisée de type Lasso pour l'analyse de données biologiques de grande dimension : application à la charge virale du VIH censurée par une limite de quantification et aux données compositionnelles du microbiote.Université de bordeauxNovember 2019
- 76 articleGene Expression Profiles Are Different in Venous and Capillary Blood: Implications for Vaccine Studies.Vaccine34442016, 5306--5313
- 77 articleJoint modelling of bivariate longitudinal data with informative dropout and left-censoring, with application to the evolution of CD4+cell count and HIV RNA viral load in response to treatment of HIV infection.Statistics in Medicine2412005, 65-82
- 78 incollectionReservoirPy: An Efficient and User-Friendly Library to Design Echo State Networks.Artificial Neural Networks and Machine Learning – ICANN 202012397ChamSpringer International Publishing2020, 494--505
- 79 articleEstimating mixed-effects differential equation models.Statistics and Computing2412014, 111--121
- 80 articleStatistical methods for HIV dynamic studies in AIDS clinical trials.Statistical Methods in Medical Research1422005, 171--192
- 81 articleATLAS: an automated association test using probabilistically linked health records with application to genetic studies.Journal of the American Medical Informatics Association2812December 2021, 2582-2592

