2024Activity reportProject-TeamCRONOS
RNSR: 202224368W- Research center Inria Centre at Université Côte d'Azur
- Team name: Computational modelling of brain dynamical networks
- Domain:Digital Health, Biology and Earth
- Theme:Computational Neuroscience and Medicine
Keywords
Computer Science and Digital Science
- A5.1.4. Brain-computer interfaces, physiological computing
- A6.1. Methods in mathematical modeling
- A9.2. Machine learning
- A9.3. Signal analysis
Other Research Topics and Application Domains
- B1. Life sciences
- B1.2. Neuroscience and cognitive science
- B1.2.1. Understanding and simulation of the brain and the nervous system
- B1.2.2. Cognitive science
- B1.2.3. Computational neurosciences
- B2.2.6. Neurodegenerative diseases
- B2.5.1. Sensorimotor disabilities
- B2.6.1. Brain imaging
1 Team members, visitors, external collaborators
Research Scientists
- Théodore Papadopoulo [Team leader, Inria, Senior Researcher, HDR]
- Rachid Deriche [Inria, Emeritus, HDR]
- Samuel Deslauriers-Gauthier [Inria, ISFP]
- Olivier Faugeras [Inria, Emeritus, HDR]
- Romain Veltz [Inria, Researcher, HDR]
Post-Doctoral Fellow
- Noémie Gonnier [Inria, Post-Doctoral Fellow, until Sep 2024]
PhD Students
- Yanis Aeschlimann [Université Côte d’Azur]
- Igor Carrara [Université Côte d’Azur, until Oct 2024]
- Laura Gee [Inria, from Dec 2024]
- Petru Isan [CHU Nice]
- Emeline Manka [Université Côte d’Azur, from Oct 2024]
Technical Staff
- Igor Carrara [Inria, Engineer, from Oct 2024 until Nov 2024]
- Laura Gee [Inria, Engineer, until Nov 2024]
- Eleonore Haupaix-Birgy [Inria, Engineer, until Sep 2024]
- Evgenia Kartsaki [Inria, Engineer, from Oct 2024]
Interns and Apprentices
- Esin Bahar Akcay [Université Côte d’Azur, Intern, from Sep 2024]
- Emeline Manka [Université Côte d’Azur, Intern, from Mar 2024 until Aug 2024]
- Teodora-Lucia Stei [Université Côte d’Azur, Intern, from Sep 2024]
Administrative Assistant
- Claire Senica [Inria]
2 Overall objectives
The objective of Cronos is to develop models, algorithms, and software to estimate, understand, and quantify the whole brain dynamics. We will achieve this objective by modeling the macroscopic architecture and connectivity of the brain at three complexity levels: source, sensor, and group (see Figure 1). These three levels, detailed in Section 3, will be studied through the unifying representation of dynamic networks.
Cronos aims at pushing forward the state-of-the-art in computational brain imaging by developing computational tools to integrate the dynamic and partial information provided by each measurement modality (fMRI, dMRI, EEG, MEG, ...)1 into a consistent global dynamic network model.
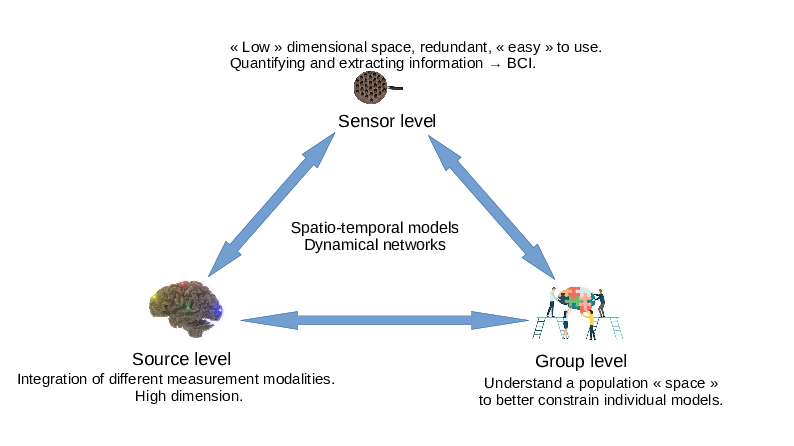
A graphic representation of the 3 Cronos axes (sensor, source and group) and their interactions.
Developing the computational models, algorithms, and software to estimate, understand, and quantify the brain's dynamical networks is a mathematical and computational challenge. Starting from several specific partial models of the brain, the overall goal is to assimilate in a single global numerical model the diversity of observations provided by non-invasive imaging. Indeed, the observations provided by various modalities are based on very different physical and biological principles. They thus reveal very different but complementary aspects of the brain, including its structural organization, electrical activity, and hemodynamic response. In addition, because of the varied nature of the physical principles used for imaging, different modalities also have drastically different spatial and temporal resolutions making their unification challenging. Some examples of sub-objectives we develop are:
- Go beyond the hypothesis that brain networks are static over a given period of time.
- Study the possibility of estimating brain functional areas simultaneously with the networks.
- Study the impact on the reconstructed networks of various models of activity transfer between brain areas.
- Develop a complete modeling of time delays involved by long range connections.
- Develop network based regularization methods in estimating brain activity.
- Go from subject specific studies to group level studies and increase the level of abstraction implied by the use of brain networks to open research perspectives in identifying network characteristics and invariances.
- Study the impact of the model imperfections on the obtained results.
Finally, the software implementing our models and algorithms must be accessible to non-technical users. They must therefore provide a level of abstraction, ease of use, and interpretability suitable for neuroscientists, cogniticians, and clinicians.
2.1 Main Research hypotheses
To achieve our goals, we decided to make some hypotheses on the way brain signals and networks are modeled.
2.1.1 Brain Signal Modeling
To model brain signals, we adopt a signal processing perspective, i.e. consider that at the macroscopic scale we look at signals, general signal modeling approaches are sufficient for the goal of describing brain dynamics. This contrasts with many other teams, which often follow a more constructivist approach, where brain signal models are derived from a computational neuroscience perspective, i.e. emerge from a mathematical modeling of neurons, axons and dendrites or assemblies of those (spiking neurons, neural mass models, ...). Our approach gives more freedom to model signals and allows for descriptions with a very limited number of parameters at the cost of losing some connection with the microscopic reality. Additionally, the reduced number of parameters simplifies the problem of parameter identification. Depending on the type of modeling, we may use rather simple phenomenological descriptions (e.g. a region is activated or not) or more or less complicated signal models (sampled signals with no specific temporal models, various multivariate autoregressive models, integro-differential equations, ...). Generally speaking, the more we will be interested in dynamic properties of the signals, the more we will need sophisticated signal models.
2.1.2 Network Modeling
Classical – deterministic – networks only strictly exist at the microscopic level where neurons are connected together through axons. For usual neural systems, such networks are huge, currently unavailable and too complex for a macroscopic view of the brain. At such a scale, networks are of stochastic nature. Both nodes and edges are only defined probabilistically. We allow ourselves to work either directly with these probablistic networks or with deterministic networks obtained by thresholding the probabilistic ones, in which case it is important to pay attention to the amount of approximation and bias introduced by the thresholding operation. Its is important to note that, while deterministic networks have received a lot of attention, the domain of stochastic networks or its tranfer to computational neurosciences is still in its infancy.
3 Research program
As described in Section 2 and Figure 1, the research program of Cronos is organized in 3 main axes corresponding to different levels of complexity:
- The Sensor level aims at extracting information directly from sensor data, without necessarily using the underlying brain anatomy. It can be thought of as a projection of the underlying brain networks onto a low dimensional space thus allowing computationally efficient processing and analysis. This provides a first window for observing and estimating brain dynamics. This sensor space is particularly convenient for the estimation of properties that are projection invariants (e.g. number of sources, state changes, ...). It is also the level that is most suitable for real time applications, such as brain-computer interfaces (BCI).
- The Source level aims at measurement integration into a unified and high dimensional spatio-temporal space, i.e. a computational representation of dynamic brain networks. This is the level that is understood by neuroscientists, cogniticians, and clinicians, and therefore has an essential role to play in the visualization of brain activities.
- The Group level aims at providing tools to constrain the search space of the two previous levels. This is the natural space to develop statistical models and tests of the functioning of the brain. It is the level that allows the study of inter-subject variability of brain activity and the development of data driven priors. In particular, the topic of making statistics over noisy networks is a challenging task for this level.
These three levels closely interact with each other, as depicted by Fig. 1, towards the ultimate goal of non-invasively and continuously localizing brain activity in the form of brain networks.
3.1 Sensor level: the first window on brain dynamics
Sensor level mostly concerns MEG and EEG measurements. These serve as a support not only for experiments aimed at understanding the functioning of the brain when it performs certain tasks, but also for the characterization of certain pathologies (such as epilepsy). As an inexpensive modality, EEG is also widely used for the development of brain-computer interfaces. EEG and MEG are characterized by a high temporal resolution (up to 1000Hz or more), by a poor spatial resolution (measurable events involve several centimers-square portions of the cortex) and by a rather poor signal-to-noise ratio (SNR)2. For both EEG and MEG, the obtained measurements are linear mixtures of the “true” electrical sources that are at a distance of the sensors. MEG and EEG temporal resolution characteristics allow them to reveal changes in the dynamic of the brain activity. This “sensor space” has two main advantages: 1) its relatively small dimension (from a few to several hundreds sensors) compared to that of the “source space” (tens of thousands of degrees of freedom) and 2) its ease-of-use3. It is thus opportune to extract as much information as possible from this lower dimensional space, which already contains all the dynamical information of the functioning brain available from these modalities. This involves a better understanding of the invariants between source and sensor levels, which can lead to better BCI classification algorithms. BCI not only clearly is an applicative domain that will benefit from this research, but will also help in validation of algorithms and methods. Our ambition is also to study how BCI-like techniques can be used for constraining the reconstruction of brain networks, for detecting some brain events in real time for clinical applications or for creating improved cognitive protocols using real time responses to dynamically adapt stimulations.
This axis relies on three major ideas, which are necessary steps towards the ultimate goal of using MEG or EEG systems to non-invasively and continuously localize brain activity in the form of brain networks (i.e. without averaging multiple trials signals).
3.1.1 Automating the detection of brain state changes from the acquired data
Experimental practice in MEG, EEG, and BCI shows that very valuable information can be obtained directly from the “sensor space”. This encompasses estimating the number 42 or the time courses of sources 36, 35 or the detection of changes in the brain dynamical activity. This type of information can be an indication of a “brain state change”. In particular, automating the detection of changes in brain states would allow the splitting of the incoming functional data into segments in which the brain network can be considered as stationary. During such stationary segments of data, the sources and their locations remain fixed, which offers a means to regularize the solution of the inverse problem of source reconstruction. Furthermore, any improvement on EEG signal classification has direct applications for BCI. One approach that we explore is the use of autoregressive spatio-temporal models or lagged correlation measurements as a means to improve classification of MEG/EEG signals. These autoregressive spatio-temporal models constitute a possible first step towards using more sophisticated causality modeling.
3.1.2 Better modeling of the spatio-temporal variability of brain signals
Hand in hand with the previous sub-goal is the need to better understand the spatio-temporal variability of brain signals. This variability originates from several factors ranging from the intrinsic variability of the brain sources (even in a same subject) to important differences in the spatial organization of the cortex across subjects or to variations in the way sensors are setup by experimenters. Thus, variability occurs not only across subjects, but also across sessions or even trials for a same subject.
The most common approach developed to cope with the noisy MEG and EEG signals is commonly referred to as evoked potentials: the signal is “clocked” on some stimulus or reaction of the subject and the low amplitude signals – almost completely hidden by background activity – are then averaged over multiple repetitions (trials) of the experiment to improve the SNR. But, because of variability, such an averaging distorts the overall activity and hides specific activities such as high frequency components. It is thus advisable to improve models so that they can “work” in single event mode (i.e. without averaging). Relying on multiple trials to obtain better statistical models of individual signals (with techniques such as dictionary learning, autoregressive models, deep-neural networks, ...) would be an important improvement over the current state-of-the-art as it would provide a more objective and systematic way of characterizing single trial data. Events will be extracted separately for each trial without relying on averaging, but with the knowledge of the “group” model (see Section 3.3). This will allow the study of the variability of brain activity across trials (attention, habituation are e.g. known to change the activity). This is a difficult long term challenge with possible short-middle term advances for some specific cases such as epileptic spikes. This research path is grounded by some of our previous work 5, 2. Understanding variability is particularly important for BCI as it is often necessary to “learn” a classifier to detect the subject's brain state with a limited dataset (because of time constraint). At the sensor level, spatial patterns of activity are often described by covariance/correlation matrices. Using Riemannian metric over the space of symmetric definite positive matrices is a powerful technique that has been used these last years by the BCI community 32. Extending and improving these techniques as well as finding proper low dimensional spaces that characterize brain activity are other research paths we will follow.
3.1.3 Adapting to new sensor modalities
During the last few years, tremendous improvements have been made on means to acquire functional brain data. Yet, novelty in this domain continues and new sensors are regularly proposed. These sensors can offer more accurate measurements with improved SNR and/or some ease-of-use improvements. For example, the use of room temperature MEG sensors (such as those developed by Mag4Health – see Section 7.1) promise improvements in both aspects. EEG dry electrodes hold the promises to simplify the setup of BCI systems, but are difficult to master. MR machines are also more and more powerful with either higher fields (better signal quality and/or better spatial resolution) or, on the contrary, lower fields (easier to use and better contrasts in some cases). It is important to continually adapt processing methods to exploit the specificities of these new sensors as they become available to the community.
3.2 Source level: the integration space
MR images of the brain offer different views of its organization and function via dMRI tractography and fMRI connectivity. When combined with MEG and EEG, we obtain complementary, but highly heterogeneous perspectives on brain networks and their dynamics. The natural space to integrate this complex information is the space of brain sources, allowing a unified model of imaging data. This axis is built over our past research (in the former team Athena) in modeling the propagation of the electromagnetic field from brain sources to sensors 6, brain structural connectivity 3 or mapping different brain imaging modalities 4. Our plan for fusing the data originating from different modalities into a single network based model can be described in 3 sub-topics.
3.2.1 Source modeling: anatomical, temporal, and numerical constraints
Brain sources refer to regions of interests whose properties link brain activity to the observed measurements. For example, in EEG, brain sources can be modeled as dipoles representing the superposition of many synchronous and parallel neurons. The combination of the electric fields generated by these dipoles gives rise to the potential differences measured at the surface of the scalp in EEG. The magnetic fields generated by these same dipoles give rise to MEG signals. Regardless of the modality, the task of recovering brain sources from imaging data is an inverse problem. This inverse problem is ill–posed, either because of the limited number of sensors (EEG and MEG), because of poor signal to noise ratio (EEG, MEG, fMRI), or because of the limited spatial (dMRI, EEG, MEG) or temporal resolution (fMRI, dMRI). To recover a unique and stable solution, simple mathematical (i.e. not fully grounded by biology) criteria such as minimum norm or sparsity are used to constrain the source space. However, these regularization approaches do not correspond to any specific anatomical or physiological properties of the brain and are therefore quite arbitrary. The challenge we address here is to increase the amount of subject specific anatomical and physiological constraints taken into account for the recovery of brain activity from non–invasive imaging. More specifically, we will investigate how brain networks, and in particular their associated delays, can be used to constrain the inverse problem. Previous work has already shown the potential of temporal regularization based on brain connectivity 3, but the topic remains largely open to identifying new models grounded in physiological data. Another difficulty is the dynamic aspect of brain networks: we first assume that a stationary brain state has been identified (e.g. using methods from Section 3.1). Eventually (as a long term objective), transitions between stationary brain state models could be directly modeled in the networks themselves and estimated from the data. The validation of the proposed models is both challenging and essential and will be explored with our clinical partners (see Section 8). Finally, non-traditional imaging modalities will also be considered. For example, accurate modeling of electromyography, in collaboration with Neurodec, can provide important timing information of sources (see Section 6.4).
3.2.2 Unified network models explaining various brain measurements
In the previous section, brain activity is estimated from one modality using information from the others as constraints or priors. This obviously favors one modality over the others and propagates errors of processing pipelines via the constraints. This modus operandi, while suboptimal, is historically justified: analysis pipelines have been developed independently for each modality by different communities. Given the complementary nature of the different modalities, it seems relevant to instead construct one global pipeline encompassing all brain measurement modalities into a single unified framework. With brain networks arising as a central concept in the neuroscientific community, we propose to make it the basis of such a global model. Doing so will enable the recovery of brain network dynamics from non–invasive multi–modal data, an important open problem in neuroscience. To achieve this objective, we propose to devise forward models describing the link between brain networks and each of the various imaging modalities. Specifically, effort is needed to evaluate different formalisms describing brain networks that differ in the way nodes and their interactions are defined. For example, we investigate the relationship between electrical (EEG/MEG), architectural (dMRI) and metabolic (fMRI) activities. Separated models – either electric or hemodynamic – have been proposed for some time, but coupling them is an important challenge all the more that the role of some neural constituents (such as glia) is currently not well understood. Such models have already been – at least partly as in 34 – devised. The main challenge is to define one that is sufficiently simple and well spatialized (in particular to incorporate connectivity) in order to be useful for our purpose. We will also consider electromyography in this context, extending brain networks to the peripherial nervous system.
3.2.3 Global inverse problem using all measurements altogether
Models established in the previous section can be used in a global inverse problem involving all the measurements modalities to recover the specific brain network involved in a task. In all cases, we need to solve an inverse problem over a complex network model with a sparse prior as we need to find “simple” networks that can explain our data. Depending on the way networks are modeled, some difficulties may arise. For example, the model defined in 40, 39 leads to combinatorial explosion when used for source localization. Finding appropriate models that facilitate the resolution of this inverse problem may require some iterations between this sub-task and the previous one.
3.3 Group level: understanding variability to constrain network properties
Given the high intra- or inter-subject variability of brain activity, it is particularly interesting to be able to characterize the part of the activity that remains invariant (over time for one subject or across different subjects). This will allow a better understanding of brain processes as well as a better understanding of their variability across time or subjects. It also opens the possibility to constrain network models to reduce their complexity and improve their identifiability. The following description mostly refers to group statistics at the source level, but similar techniques are also relevant for the sensor level axis (see Section 3.1). This axis is a long term research effort as it builds upon previous axes.
3.3.1 Matching individual subject models
Reconstructions obtained in Section 3.2 will certainly differ even for different measurement sessions obtained with a same subject. An important problem to solve is the matching of instances of such reconstructions (whether they consist in brain networks or spatio-temporal autoregressive models). In general, the relative positions of functional brain areas are fairly well known. However, temporal variations (lag, duration) in these models make the matching problem rather complicated. Finding and using some invariants of the models is one path to find a good compromise that would allow for enough flexibility in the matching process while avoiding the combinatorial complexity that e.g. graph matching problems usually exhibit. Another – long term – possibility would be to directly solve “group problems”, but this would complexify even more the modeling problem and might have the same drawbacks as averaging if not properly done (i.e. hiding some activity or the intrinsic variability of the studied phenomenon).
3.3.2 Statistics over brain networks
Once the matching of single subject models has been solved, statistics will be necessary to assess the significance of the various model elements (e.g. in the case of dynamic network models, brain areas and their connections). Such statistics can also be used as a basis to derive “group statistical models” describing a family of task-related models that can in turn be used to constrain models used in Section 3.2 and, through a forward model to reduce the dimension of the search space at sensor level (see Section 3.1). The BCI community is still divided on the actual benefits in terms of accuracy on using the source space (v.s. the sensor space) for classifying brain signals. The statistical tools developed in this sub-axis may help to address this problem.
4 Application domains
4.1 Clinical applications
Cronos research has a strong clinical potential impact for brain diseases like epilepsy, brain cancer surgery, phantom pains, traumatic brain injuries, ...We closely work with several hospital teams and research groups (Pasteur hospital in Nice, La Timone hospital in Marseille, CRNL – Centre de Recherche en Neuroscience in Lyon) towards exporting our research in their medical contexts. Example of applications are:
- Better understanding brain stimulation used in awake brain surgery and its relation with brain anatomy and in particular fibers as measured by dMRI.
- Real time detection of epileptic spikes from new generation MEG data and the visualization of their associated brain sources in real time.
- Use of BCI (see Section 4.2) for helping disabled people to communicate (e.g. for patients suffering from Amyotrophic Lateral Sclerosis) or for helping epileptic patients to learn how to control the occurence of seizures.
- Understand the somatotopy of the thalamus and its relation with the efficiency of the deep brain stimulation therapy.
4.2 Brain Computer Interfaces (BCI)
BCI is a closely related domain to both the “Signal Processing” (Section 2.1.1) and “Network Modeling” (Section 2.1.2) aspects. It typically extracts from the signal a “brain state”' that is a correlate of the “brain network”. While traditional MEG/EEG studies have been extensively exploited by the BCI community, the opposite nourishing of the former field by BCI has been much less explored. There is a continuing dispute in the BCI community on the advantages of going to source space or not (see e.g. 41). By studying the invariants between source and sensor space, we hope not only to provide clues on the above dispute but also to open the opportunity of using BCI-like techniques to ease the more traditional brain signal processing. In some sense, such invariants are the information that BCI exploits in doing classification on sensor data. BCI also has the advantage of providing a more quantitative way (in term of classification quality) to evaluate methods. Controlling the amount of resources (computer power, number and quality of the sensors, ...) needed to achieve a given classification accuracy is also a strong BCI concern. This point of view is also complementary to that of more traditional brain signal modeling and can have a significant impact in terms of cost and ease of use in the clinical context.
5 New software, platforms, open data
5.1 New software
5.1.1 BCI-VIZAPP
-
Name:
Real time EEG applications
-
Keyword:
EEG
-
Functional Description:
BCI-VIZAPP is a software suite for designing real-time EEG applications such as BCIs or neurofeedback applications. It has been been developed to build a virtual keyboard for typing text and a photodiode monitoring application for checking timing issues, but can now be also used in other tasks such as EEG monitoring. Originally, it was designed to delegate signal acquisition and processing to OpenViBE but has recently been extended to get some of these capabilities. This allows for more integrated and robust applications but also opens up new algorithmic opportunities, such as real time parameter modification, more controlled interfaces, ...
-
News of the Year:
Bci-Vizapp has been enriched with numerous features, notably to read and save certain files created with OpenViBE or with MNE-python, thus allowing an easier communication with them. Bci-Vizapp is also the software base used (or which we aim to use it in the long term) for different contracts (Demagus, EPIFEED, EPINFB, Techicopa and ConnectTC) and has integrated different elements to support them.
-
Contact:
Théodore Papadopoulo
-
Participants:
Nathanael Foy, Romain Lacroix, Maureen Clerc Gallagher, Théodore Papadopoulo, Yang Ji, Come Le Breton
5.1.2 BifurcationKit
-
Name:
Automatic computation of numerical bifurcation diagrams
-
Keywords:
Bifurcation, GPU
-
Functional Description:
This Julia package aims at performing automatic bifurcation analysis of possibly large dimensional equations function of a real parameter by taking advantage of iterative methods, dense / sparse formulation and specific hardware (e.g. GPU).
It incorporates continuation algorithms (PALC, deflated continuation, ...) based on a Newton-Krylov method to correct the predictor step and a Matrix-Free/Dense/Sparse eigensolver is used to compute stability and bifurcation points.
The package can also seek for periodic orbits of Cauchy problems. It is by now one of the few software programs which provide shooting methods and methods based on finite differences or collocation to compute periodic orbits.
The current package focuses on large-scale, multi-hardware nonlinear problems and is able to use both Matrix and Matrix Free methods on GPU.
-
News of the Year:
The package performance (4x faster) was improved with a new method for solving the linear problem of the collocation approximation of periodic orbits. A new plotting backend for Makie.jl has been added. Additionally, a package MultiParamContinuation.jl has been added to the github organisation bifurcationkit. It allows computations of zeros of functional as function of two parameters instead of one in BifurcationKit.jl.
- URL:
- Publication:
-
Contact:
Romain Veltz
-
Participant:
Romain Veltz
6 New results
6.1 Sensor Level: Brain Signal Modeling (see Section 3.1)
Augmented Covariance Approaches for BCI
Participants: Igor Carrara, Théodore Papadopoulo.
EEG signals are complex and difficult to characterize, in particular because of their variability. They therefore require the use of specific and adapted signal processing methods. In a recent approach 33, sources were modeled as an autoregressive model which explains a portion of a signal. This approach works at the level of the source space (i.e. the cortex), which requires modeling of the head and makes it quite expensive. However, EEG measurements can be considered as a linear mixture of sources and therefore it is possible to estimate an autoregressive model directly at the measurement level. The objectives of this line of work is to explore the possibility of exploiting EEG/MEG autoregressive models to extract as much information as possible without requiring the complex head modeling needed for source reconstruction.
A first result in this line of research is the use of Augmented Covariance Matrices (ACMs) for BCI classification. These ACMs appear naturally in the Yule-Walker equations that were derived to recover autoregressive models from data. ACMs being symmetric positive definite (SPD) matrices, it is natural to apply Riemannian geometry based classification approaches on these objects as they represent the current state-of-the-art. Using these ACMs noticeably improves classification performance on several BCI benchmarks both in the within-session or in the cross-session evaluation protocols4. This is due to the fact that ACMs incorporate not only spatial (the classical covariance matrix) but also temporal information. As such, it contains information on the nonlinear components of the signal through the embedding procedure, which allows the leveraging of dynamical systems algorithms. This comes at the cost of introducing two hyper-parameters: the order of the autoregressive model and a delay that controls the time resolution at which the signal is considered, which have to be estimated.
The improvements related above are made with a grid search hyper-parameter selection, which can be quite costly. As an ACM can be seen as the standard spatial covariance matrix of an extended signal created from the original one using a multidimensional embedding, there is an obvious link with the delay embedding theorem proposed by Takens 44 for the study of dynamical systems. This view provides an alternate path for estimating the hyper-parameters using methods proposed for dynamical systems such as the MDOP (Maximizing Derivatives On Projection) algorithm 43.
This work is described in the article 10.
Block Toeplitz Augmented Covariance Approaches for BCI
Participants: Igor Carrara, Théodore Papadopoulo.
As explained above, standard ACM emerges as a combination of phase space reconstruction of dynamical systems and of Riemannian geometry. This approach creates a matrix that contains not only an average spatial representation of the signal but also a representation of its evolution in time. In addition to be SPD, ACM matrices also possesses the structural property of being Block-Toeplitz (BT), which remained un-exploited. Recently, an approach has been proposed to better deal with such BT SPD matrices in the fields of audio processing or radar signal analysis. The idea of this research is thus to endow the smooth manifold of BT SPD matrices with a Riemannian metric, thus allowing the ACM matrix to be treated within its true manifold membership. For this, we use a mapping from the BT SPD matrix manifold to the product of an SPD manifold and several Siegel disk spaces, which is obtained by applying an appropriate conversion of the blocks of the ACM matrix into the Verblusky coefficients. The new – Siegel metric based – pipeline was tested and validated against several state-of-the-art Machine Learning (ML) and Deep Learning (DL) algorithms on several datasets for motor imagery (MI) classification using several subjects and several tasks using the MOABB (Mother Of All BCI Benchmark) framework, and a within-session evaluation procedure. The resulting algorithm achieves a performance that is significantly better than state-of-the-art BCI algorithms, except for TS+EL (tangent space Riemannian metric with elastic-net classification) to which it is comparable. However, this novel algorithm achieves this performance with a substantial reduction in computational costs and carbon footprint compared with standard ACM.
This work is described in the preprint 23 and has been presented in the conferences CORTICO, MicroStates and Graz BCI 17, 31, 30.
Phase-SPDNet: a deep learning network based on the augmented covariance framework
Participants: Igor Carrara, Bruno Aristimunha [Universidade Federal do ABC, Santo André, Brazil and Université Paris-Saclay, Paris, France], Marie-Constance Corsi [NERV, Inria, Paris Brain Institute, France], Raphael Yokoingawa De Camargo [Universidade Federal do ABC, Santo André, Brazil], Sylvain Chevallier [Université Paris-Saclay, Paris, France], Théodore Papadopoulo.
The integration of DL algorithms on brain signal analysis is still in its nascent stages compared to their success in fields like Computer Vision. This is particularly true for BCI, where the brain activity is decoded to control external devices without requiring muscle control. EEG, while a widely adopted choice for designing BCI systems due to its non-invasive, cost-effectiveness and excellent temporal resolution, comes at the expense of limited training data, poor signal-to-noise ratio, and a large variability across and within-subject recordings. Furthermore, setting up a BCI system with many electrodes takes a long time, hindering the widespread adoption of BCIs outside research laboratories. To boost adoption, we need to improve user comfort using reliable algorithms that operate with few electrodes. Our research aimed at developing a DL algorithm that delivers effective results with a limited number of electrodes. Taking advantage of the ACM and the framework of SPDNet 37, we propose the Phase-SPDNet architecture and analyze its performance and the interpretability of the results. The evaluation is conducted on 5-fold cross-validation, using only three electrodes positioned above the motor cortex. The methodology was tested on nearly 100 subjects from several open-source datasets using the MOABB framework. The augmented approach combined with the SPDNet architecture significantly outperforms all the current state-of-the-art DL architecture in MI decoding. Furthermore, this new architecture is explainable and has a low number of trainable parameters.
This work has been published in 9.
Pseudo-online framework for BCI evaluation
Participants: Igor Carrara, Théodore Papadopoulo.
BCI technology operates in three modes: online, offline, and pseudo-online. In the online mode, real-time EEG data is constantly analyzed. In offline mode, the signal is acquired and processed afterwards. The pseudo-online mode processes the collected data as if they were received in real-time, but without the real-time constraints. The main difference is that the offline mode often analyzes the whole data, while the online and pseudo-online modes only analyze data in short time windows, which is more flexible and realistic for real-life applications. Consequently, offline processing tends to be more accurate, but biases the comparison of classification methods towards methods requiring more dataset. In this work, we extended the current MOABB framework 38, that used to operate in offline mode only, with pseudo-online capabilities using overlapping sliding windows. This requires the introduction of an idle state event in the dataset to account for non-task-related signal portions. We analyzed the state-of-the-art algorithms of the last 15 years over several MI datasets composed by several subjects, showing the differences between the two approaches (offline, and pseudo-online) and demonstrate that the adoption of the pseudo-online strategy modifies the hierarchy of classification methods.
This work has been published in 11.
The largest EEG-based BCI reproducibility study for open science: the MOABB benchmark
Participants: Sylvain Chevallier [Université Paris-Saclay, Paris, France], Igor Carrara, Bruno Aristimunha [Universidade Federal do ABC, Santo André, Brazil and Université Paris-Saclay, Paris, France], Pierre Guetschel [Donders Institute for Brain, Cognition and Behaviour], Sara Sedlar [Université Paris-Saclay], Bruna Junqueira Lopes [ISAE-SUPAERO - Institut Supérieur de l'Aéronautique et de l'Espace], Sébastien Velut [ISAE-SUPAERO - Institut Supérieur de l'Aéronautique et de l'Espace and Université Paris-Saclay, Paris, France], Salim Khazem [Université Paris-Saclay, Paris, France], Thomas Moreau [Neurospin CEA and Inria Saclay - Ile de France].
This study conducts an extensive BCI reproducibility analysis on open EEG datasets, aiming to assess existing solutions and establish open and reproducible benchmarks for effective comparison within the field. The need for such benchmarks lies in the rapid industrial progress that has given rise to undisclosed proprietary solutions. Furthermore, the scientific literature is dense, often featuring challenging-to-reproduce evaluations, making comparisons between existing approaches arduous. Within an open framework, 30 ML pipelines (separated into raw signal: 11, Riemannian: 13, DL: 6) are meticulously re-implemented and evaluated across 36 publicly available datasets, including MI (14), P300 (15), and SSVEP (7)5. The analysis incorporates statistical meta-analysis techniques for result assessment, encompassing execution time and environmental impact considerations. It yields principled and robust results applicable to various BCI paradigms, emphasizing MI, P300, and SSVEP. Notably, Riemannian approaches utilizing spatial covariance matrices exhibit superior performance, underscoring the necessity for significant data volumes to achieve competitive outcomes with DL techniques. The comprehensive results are openly accessible, paving the way for future research to further enhance reproducibility in the BCI domain. The significance of this study lies in its contribution to establishing a rigorous and transparent benchmark for BCI research, offering insights into optimal methodologies and highlighting the importance of reproducibility in driving advancements within the field.
This work has been submitted for publication and is currently available as the preprint 24.
6.2 Source Level: Brain Dynamic Network Modeling (see Section 3.2)
On null models for temporal small-worldness in brain dynamics
Participants: Aurora Rossi [COATI, Inria], Samuel Deslauriers-Gauthier, Emanuele Natale [COATI, Inria].
Progress was made in modeling brain dynamics as temporal networks using fMRI data. A key focus was on developing statistical null models to support hypothesis validation in temporal network research. To address this, the Random Temporal Hyperbolic Graph (RTH) model was introduced, building on the established Random Hyperbolic Graph (RH) model, which is often used to describe properties of real-world networks. The study examined temporal small-worldness, a property associated with efficient information exchanges in brain networks and well-documented in static networks. Results indicated that the RTH model more accurately reflects the small-worldness of resting-state brain activity compared to conventional null models for temporal networks. Notably, the RTH model achieves this with minimal additional complexity, requiring only one additional parameter compared to classical approaches. These findings suggest that the RTH model may offer a useful framework for studying brain network dynamics and validating hypotheses in temporal network analysis.
This work has been published in 15.
Brain surgery: The effect of common parameters of bipolar stimulation on brain evoked potentials
Participants: Petru Isan [Pasteur Hospital, UR2CA, Nice], Samuel Deslauriers-Gauthier, Théodore Papadopoulo, Denys Fontaine [Pasteur Hospital, UR2CA, Nice], Patryk Filipiak [NYU Langone Health], Fabien Almairac [Pasteur Hospital, UR2CA, Nice].
Bipolar direct electrical stimulation (DES) of an awake patient is the reference technique for identifying brain structures to achieve maximal safe tumor resection. Unfortunately, DES cannot be performed in all cases. Alternative surgical tools are, therefore, needed to aid identification of subcortical connectivity during brain tumor removal. In this study, we identified optimal bipolar stimulation parameters for robust generation of brain evoked potentials (BEPs), namely the interelectrode distance (IED) and the intensity of stimulation (IS), in cortical and axonal stimulation. BEPs were elicited using bipolar stimulation under varying IEDs and ISs. Results indicated that increasing IS led to higher amplitudes of the N1 wave, a key electrophysiological marker of cortical and subcortical activity. Additionally, axonal stimulation produced more robust N1 responses with shorter latencies compared to cortical stimulation, while peak delays remained unaffected by the stimulation parameters. These findings provide insights into optimizing stimulation settings for effective brain mapping. The study highlights the importance of understanding how stimulation parameters influence BEPs, particularly the N1 wave, which reflects structural and functional connectivity. By refining these parameters, we aim to improve the reliability of BEPs in identifying cortical and white matter structures during surgery.
This work has been published in 13
Brain subnetwork analysis in narrative data
Participants: Aurora Rossi [COATI, Inria], Yanis Aeschlimann, Emanuele Natale [COATI, Inria], Peter Ford Dominey [CNRS], Samuel Deslauriers-Gauthier.
Functional connectivity derived from functional Magnetic Resonance Imaging (fMRI) data has been increasingly used to study brain activity. In this work, we model brain dynamic functional connectivity during narrative tasks as a temporal brain network and employ a ML model to classify in a supervised setting the modality (audio, movie), the content (airport, restaurant situations) of narratives, and both combined. Leveraging Shapley values, we analyze subnetwork contributions within Yeo parcellations (7- and 17-subnetworks) to explore their involvement in narrative modality and comprehension. This work represents the first application of this approach to functional aspects of the brain, validated by existing literature, and provides novel insights at the whole-brain level. Our findings suggest that schematic representations in narratives may not depend solely on pre-existing knowledge of the top-down process to guide perception and understanding, but may also emerge from a bottom-up process driven by the ventral attention subnetwork.
Connectivity in the rat brain
Participants: Samuel Deslauriers-Gauthier, Joana Gomes-Ribeiro [Instituto de Medicina Molecular, Portugal], João Martins [Instituto de Ciências Nucleares Aplicadas à Saúde and University of Coimbra, Portual], Teresa Summavielle [Instituto de Investigação e Inovação em Saúde, Portugal], Joana E. Coelho [Instituto de Medicina Molecular, Portugal], Miguel Remondes [Instituto de Medicina Molecular, Portugal], Miguel Castelo Branco [Instituto de Ciências Nucleares Aplicadas à Sa'de and University of Coimbra, Portual], Luísa V. Lopes [Instituto de Medicina Molecular, Portugal].
Addiction to psychoactive substances is a maladaptive learned behavior. Contexts surrounding drug use integrate this aberrant mnemonic process and hold strong relapse-triggering ability. In 12, we asked where context and salience might be concurrently represented in the brain during retrieval of drug-context paired associations. For this, we developed a morphine-conditioned place preference protocol that allows contextual stimuli presentation inside a MRI scanner and investigated differences in activity and connectivity at context recall. We found context-specific responses to stimulus onset in multiple brain regions, namely, limbic, sensory and striatal. Differences in functional interconnectivity were found among amygdala, lateral habenula, and lateral septum. We also investigated alterations to resting-state functional connectivity and found increased centrality of the lateral septum in a proposed limbic network, as well as increased functional connectivity of the “lateral habenula” and hippocampal “cornu ammonis 1” region, after a protocol of associative drug-context. Finally, we found that pre-conditioned place preference resting-state connectivity of the lateral habenula and amygdala was predictive of inter-individual score differences. Overall, our findings show that drug and saline-paired contexts establish distinct memory traces in overlapping functional brain microcircuits and that intrinsic connectivity of the habenula, septum, and amygdala likely underlies the individual maladaptive contextual learning to opioid exposure. We have identified functional maps of acquisition and retrieval of drug-related memory that may support the relapse-triggering ability of opioid-associated sensory and contextual cues. These findings may clarify the inter-individual sensitivity and vulnerability seen in addiction to opioids found in humans.
The tractography equation
Participants: Samuel Deslauriers-Gauthier, Romain Veltz.
Current dMRI fiber tractography algorithms are numerical methods that approximate the solution to an ill-defined problem. This lack of formal definition is unsurprising, given the immense appeal of estimating structural brain connectivity in vivo and non-invasively compared to the theoretical underpinning of the problem. As a result, efforts have predominantly been centered on assessing and improving tractography’s capacity to trace the trajectories of white matter bundles, rather than on its mathematical formulation or numerical precision. In this project, we mathematically investigate the tractography algorithms, and reformulate them into classical stochastic differential equations and partial differential equation. We also provide a numerical implementation of these algorithms that will be bundled in a public Python package.
6.3 Group Level (see Section 3.3)
Alignment of brain networks
Participants: Yanis Aeschlimann, Samuel Deslauriers-Gauthier, Théodore Papadopoulo, Anna Calissano [Inria EPIONE], Xavier Pennec [Inria EPIONE].
Every brain is unique, having its structural and functional organization shaped by both genetic and environmental factors over the course of its development. Brain image studies tend to produce results by averaging across a group of subjects, under a common assumption that it is possible to subdivide the cortex into homogeneous areas while maintaining a correspondence across subjects. This project questions such an assumption: can the structural and functional properties of a specific region of an atlas be assumed to be the same across subjects? In this work, this question is addressed by looking at the network representation of the brain, with nodes corresponding to brain regions and edges to their structural relationships. We perform graph matching on a set of control patients and on parcellations of different granularity to understand which is the connectivity misalignment between regions. The graph matching is unsupervised and reveals interesting insight on local misalignment of brain regions across subjects.
This work has been published in the article 8.
Structural connectivity is one view of brain networks provided by dMRI, but these networks can also be observed from a functional point of view via fMRI. We thus propose to simultaneously exploit structural and functional information in the alignment process. This allows us to explore multiple perspective of brain networks. Our results show that the permutations induced by one type of connectivity (structural, functional) are not always supported by the other connectivity network, but when considering a combined alignment, it is possible to find permutations of regions which are supported by both connectome modalities, leading to an increased similarity of functional and structural connectivity across subjects.
6.4 Other Results
CNN interpretability for self-supervised models
Participants: Aymene Mohammed Bouayed [Be-Ys Research, France], Samuel Deslauriers-Gauthier, Adrian Iaccovelli [Be-Ys Research, France], David Naccache [DIENS, ENS, CNRS, PSL University, Paris, France].
Interpreting the decisions of Convolutional Neural Networks (CNNs) is essential for understanding their behavior, yet it remains a significant challenge, particularly for self-supervised models. Most existing methods for generating saliency maps rely on reference labels, restricting their use to supervised tasks. The EigenCAM approach is the only notable label-independent alternative, leveraging Singular Value Decomposition to generate saliency maps applicable across CNN models, but it does not fully exploit the tensorial structure of feature maps. In this work, we introduce the Tucker Saliency Map (TSM) method, which applies Tucker tensor decomposition to better capture the inherent structure of feature maps, producing more accurate singular vectors and values. These are used to generate high-fidelity saliency maps, effectively highlighting objects of interest in the input. We further extend EigenCAM and TSM into multivector variants—Multivec-EigenCAM and Multivector Tucker Saliency Maps (MTSM)—which utilize all singular vectors and values, further improving saliency map quality. Quantitative evaluations on supervised classification models demonstrate that TSM, Multivec-EigenCAM, and MTSM achieve competitive performance with label-dependent methods. Moreover, TSM enhances interpretability by approximately 50% over EigenCAM for both supervised and self-supervised models. Multivec-EigenCAM and MTSM further advance state-of-the-art interpretability performance on self-supervised models, with MTSM achieving the best results.
This work has been published in 22.
Realistic simulation of Electromyogram (EMG) data
Participants: Shihan Ma [Imperial College London, United Kingdom], Samuel Deslauriers-Gauthier, Xinjun Sheng [Shangaï Jiao Tong University, China], Xiangyan Zhu [Shangaï Jiao Tong University, China], Kostiantyn Maksymenko [Neurodec, Sophia Antipolis, France], Alexander Kenneth Clarke [Imperial College London, United Kingdom], Irene Mendez Guerra [Imperial College London, United Kingdom], Dario Farina [Imperial College London, United Kingdom].
In a previous work, we introduced the concept of a myoelectric digital twin: a highly realistic and fast computational model tailored for the training of DL algorithms. It enables the simulation of arbitrary large and perfectly annotated datasets of realistic EMG signals, allowing new approaches to muscular signal decoding, accelerating the development of human-machine interfaces.
While the myoelectric digital twin is highly accurate, it is computationally expensive and thus is generally limited to modeling static systems such as isometrically contracting limbs. As a solution to this problem, we propose a transfer learning approach, in which a conditional generative model is trained to mimic the output of an advanced numerical model. To this end, we present BioMime, a conditional generative neural network trained adversarially to generate motor unit activation potential waveforms under a wide variety of volume conductor parameters. We demonstrate the ability of such a model to predictively interpolate between a much smaller number of numerical model's outputs with a high accuracy. Consequently, the computational load is dramatically reduced, which allows the rapid simulation of EMG signals during truly dynamic and naturalistic movements.
This work has been published in 27.
Together, the myoelectric digital twin and BioMime provide a fast and accurate simulation of electromyography. However, the input to these systems is abstract: a percentage of voluntary contraction per muscle. A more intuitive and natural input would be a movement description, from which muscle contractions could be extracted. To this end, we propose NeuroMotion, an open-source simulator that provides a full-spectrum synthesis of EMG signals during voluntary movements. NeuroMotion is comprised of three modules. The first module is an upper-limb musculoskeletal model with OpenSim API to estimate the muscle fibre lengths and muscle activations during movements. The second module is BioMime, a deep neural network-based EMG generator that receives nonstationary physiological parameter inputs, such as muscle fibre lengths, and efficiently outputs motor unit action potentials. The third module is a motor unit pool model that transforms the muscle activations into discharge timings of motor units. The discharge timings are convolved with the output of BioMime to simulate EMG signals during the movement. NeuroMotion is the first full-spectrum EMG generative model to simulate human forearm electrophysiology during voluntary hand, wrist, and forearm movements.
This work has been published in 14.
Analysis of large size networks of Hopfield neurones
Participants: Olivier Faugeras, Etienne Tanré [LJAD, Nice, France].
We revisit the problem of characterizing the thermodynamic limit of a fully connected network of Hopfield-like neurons. Our contributions are: a) a complete description of the mean-field equations as a set of stochastic differential equations depending on a mean and covariance functions, b) a provably convergent method for estimating these functions, and c) numerical results of this estimation as well as examples of the resulting dynamics. The mathematical tools are the theory of Large Deviations, Itô stochastic calculus, and the theory of Volterra equations.
This work has been submitted to a journal 25 for publication.
Pros and cons of mean field representations of large size networks of spiking neurons
Participants: Olivier Faugeras, Romain Veltz.
We have laid out a roadmap for classifying the behaviors of large ensembles of networks of spiking neurons through the analysis of the corresponding nonlinear Fokker-Planck equation. Rather than using this equation to simulate the network equations, we apply advanced methods for analyzing the bifurcations of its solutions. This analysis can then be used to predict the behaviors of the original network. We have used the example of a fully connected network of Fitzhugh-Nagumo neurons with electrical and chemical synapses to convey the interest of our approach.
The work has been presented at the 2024 International Conference on Mathematical Neuroscience and a manuscript is being prepared for submission to a journal.
Topos theory and networks of neurons
Participants: Olivier Faugeras.
We have started the exploration of the application of Grothendieck's topos theory to the representation of networks of neurons. We study the work of Olivia Caramello at the University of Insubria, Italy, as well as that of Laurent Lafforgue's group at Huawei Technologies in Boulogne-Billancourt.
Excitable response of a noisy adaptive network of spiking lasers
Participants: Stéphane Barland [CNRS, Nice, France], Otti D'Huys [Maastricht University], Romain Veltz.
In the publication 21, we experimentally and theoretically analyze the response of a network of spiking nodes to external perturbations. We relate the excitable response of the network to the existence of a separatrix in phase space and analyze the effect of noise close to this separatrix. We numerically find that larger networks are more robust to uncorrelated noise sources in the nodes than small networks, in contrast to the experimental observations. We remove this discrepancy considering the impact of a global noise term in the adaptive coupling signal and discuss our observations in relation to the network structure.
Theoretical / numerical study of modulated traveling waves in inhibition stabilized networks
Participants: Safaa Habib [Université Côte d'Azur], Romain Veltz.
In the publication 26, we prove a principle of linearized stability for traveling wave solutions to neural field equations posed on the real line. Additionally, we provide the existence of a finite dimensional invariant center manifold close to a traveling wave, which allows to study bifurcations of traveling waves. Finally, the spectral properties of the modulated traveling waves are investigated. Numerical schemes for the computation of modulated traveling waves are provided. We then apply these results and methods to study a neural field model in an inhibitory stabilized regime. We showcase Fold, Hopf and Bodgdanov-Takens bifurcations of traveling pulses. Additionally, we continue the study of the modulated traveling pulses as function of the time scale ratio of the two neural populations and show numerical evidences for snaking of modulated traveling pulses.
7 Bilateral contracts and grants with industry
7.1 Bilateral Grants with Industry
Demagus: BPI Grant April 2022–March 2025
Participants: Laura Gee, Éléonore Haupaix-Birgy, Noémie Gonnier, Côme Le Breton, Théodore Papadopoulo, Julien Wintz.
This grant has been obtained with the startup Mag4Health, CNRS and INSERM (see Section 3.1.3). This company develops a new MEG machine working with optically pumped magnetometers (magnetic sensors), which potentially means lower costs and better measurements. Cronos is in charge of developing a real time interface for signal visualization, source reconstruction and epileptic spikes detection. The initial funding was for 2 years, but two extensions of 6 months have been obtained. This work is related to the research goals described in Section 3.1.3.
8 Partnerships and cooperations
8.1 International initiatives
8.1.1 Associate Teams in the framework of an Inria International Lab or in the framework of an Inria International Program
During its second year, the associated team MUSCULAR (Inria London Programme) completed work on the development of tools for rapid forward modeling in EMG (see Section 6.4). Two previously submitted preprints have now been published: one in IEEE Transactions on Neural Networks and Learning Systems 27 and another in PLOS Computational Biology 14. These advancements enable the rapid generation of realistic EMG data, a crucial capability for testing algorithms slated for development in the second and third years of the project.
As planned for the second year, the team has also made progress on its second objective: developing novel algorithms to estimate motor unit spike trains from surface EMG. Two different approaches are pursued in parallel: electrode location optimization and physics-informed inverse problems.
Electrode optimization
The first approach is based on the idea that the electrode configuration is not optimized and leads to variations in sensitivity across populations. For example, the number of identified motor neurons identified is typically larger in males than females. We also observe variations in sensitivity across target muscle groups within a single individual. Using the forward models developed during the first year, we will generate highly realistic EMG signals representing a large population of individuals with varying anatomical features. The large amount of data generated will allow us to globally optimize electrode design for specific populations (females/males) or for specific muscle architectures (e.g. pennate v.s. fusiform). The strength of this approach is to leverage highly realistic simulations to optimize over a large population whose EMG signals would be unfeasible to acquire in vivo (potentially thousands of individuals).
Physics informed inverse problems
The second approach leverages volume conductor knowledge to enhance the estimation of motor unit spike trains. Using anatomical MRI data of the forearm — acquired in various positions during the first year with optimized sequences — the team has constructed physically realistic forward models for EMG in two subjects. We are now investigating the theoretical aspects of the problem, building on our expertise in electroencephalography (EEG). One of the challenges we are currently investigating is the inclusion of the known temporal components of EMG, which are typically unknown in EEG.
8.2 International research visitors
8.2.1 Visits of international scientists
- Erol Gelenbe (Institute of Theoretical and Applied Informatics, Polish Academy of Science) visited the team in February to discuss spiking models.
- Maxime Descoteaux (Université de Sherbrooke, Canada), François Rheault (Université de Sherbrooke, Canada), Joël Lefebvre (Université du Québec à Montréal, Canada), and Laurent Petit (Université de Bordeaux, France) visited the team in March to discuss various collaboration opportunities.
- Peter-Ford Dominey (Inserm, Dijon, France) visited the team in December and gave a talk entitled “How input structure can interact with anatomy to shape functional connectivity: Theory”.
8.3 National initiatives
ANR ChaMaNe, Mathematical Challenges in Neurosciences
Participants: Romain Veltz.
- Coordinator Name :
-
Partner Institutions:
- Partner 1: Biologie Computationnelle et Quantitative (CQB), France
- Partner 2: LJAD, Université Nice, France
-
Date/Duration:
2020-2025
- Web site
This project aims at a mathematical study, on the one hand, of the intrinsic dynamics of a neuron and their consequences, and on the other hand, of the qualitative dynamics of large neural networks with respect to the intrinsic behavior of the individual neurons, the interactions between them, memory effects, spatial structure, etc.
FHU InnovPain 2
Participants: Théodore Papadopoulo.
-
Coordinator Name :
Denys Fontaine (Pasteur Hospital)
-
Date/Duration:
2023–2027
- Web site
The FHU INOVPAIN is a Hospital-University Federation focused on chronic refractory pain and innovative therapeutic solutions. It involves 6 hospital facilities, 13 academic research teams and 12 platforms and core facilities.
Inria-Inserm fellowship
Participants: Laura Gee, Théodore Papadopoulo, Christian Bénar.
-
Date/Duration:
2024–2027
The Ph.D. of L. Gee is funded by an Inria-Inserm fellowship.
ANR NeuroMotor
Participants: Samuel Deslauriers-Gauthier.
-
Coordinator Name :
François Hug (LAMHESS, Université Côte d'Azur)
-
Date/Duration:
2024–2027
There are many physical disabilities with a neurological origin, such as stroke and spinal cord injuries, that significantly impact a person’s mobility, physical capacity, stamina, or dexterity. In the era of personalized medicine, optimizing the treatment of these physical disabilities requires: i) accessing direct information on the neural commands that are sent to the muscles, and ii) integrating this knowledge into the development and assessment of rehabilitation and neurotechnology aimed at restoring movement. The breakthrough of our approach lies in changing the level at which we observe the control of movement, i.e., shifting focus from the level of whole muscles to the spinal (alpha) motor neurons. To achieve this, we will combine the use of dense grids of surface EMG electrodes with algorithms that decode the firing activity of spinal motor neurons. By unravelling the “neural code” for movement generation, we will address critical gaps in our understanding of the control of movement in health and disease. In this project, we will decode the activity of large populations of motor neurons from different muscles. We will identify motor neuron synergies, defined as functional groups of motor neurons that share inputs from various supraspinal, spinal and sensory sources. In this translational project, we will examine the structure and plasticity of these synergies in both healthy controls and patients with neurological impairments (stroke, spinal cord injury). We will also enhance the electrode design to facilitate the transfer of these methods into clinical settings.
9 Dissemination
Participants: Rachid Deriche, Samuel Deslauriers-Gauthier, Olivier Faugeras, Théodore Papadopoulo, Romain Veltz.
9.1 Promoting scientific activities
Reviewer
- T. Papadopoulo served as reviewer for the conferences ISBI and CORTICO6.
- S. Deslauriers-Gauthier served as reviewer for CVPR and ISBI.
9.1.1 Journal
Member of the editorial boards
- T. Papadopoulo serves as Associate Editor in “Frontiers: Brain Imaging Methods” and as Review Editor for “Frontiers: Artificial Intelligence in Radiology”.
- S. Deslauriers-Gauthier serves as Review Editor for “Frontiers in Computational Neuroscience”.
- R. Veltz is an Editor for “Mathematical Neuroscience and Applications”.
Reviewer - reviewing activities
- T. Papadopoulo served as reviewer for several international journals (“IEEE Transactions on Biomedical Engineering”, “IEEE Transactions on Neural Systems and Rehabilitation Engineering”, “PLOS One”, “Frontiers in Neurosciences”, “Brain Topography”, “Computers in Biology and Medicine”, “IEEE Transactions on Medical Imaging”).
- S. Deslauriers-Gauthier served as reviewer for several international journals (“Medical Image Analysis”, “Journal of Electromyography and Kinesiology”).
- R. Veltz served as reviewer for several international journals (“PLOS Computational Biology”, “Theory in Biosciences”').
9.1.2 Invited talks
- R. Veltz. Some recent results on mean fields of networks of spiking neurons, Seminar of Inria team McTao, 2024.
- S. Deslauriers-Gauthier. Modeling the Brain Connectome, AI Challenges for European Research and Academia (Academia Europaea), London, September 2024.
9.1.3 Leadership within the scientific community
- O. Faugeras is a member of the French Academy of Sciences and of the French Academy of Technologies. As such, he participates in several activities that have a significant impact on the Institute and the French scientific community. The preparation of application files for candidates to elections at the Academy and to some of the prestigious prizes which are awarded every year are two examples.
- R. Deriche is a member of Academia Europaea.
9.1.4 Research administration
- T. Papadopoulo has been named adjoint director of the NeuroMod Institute. He was in effect assuming the role of director (January-June) after the resignation of the former direction. He has then been elected in July at this same position for 4 years.
- T. Papadopoulo was the head of the Technological Development Committee of Inria Centre at Université Côte d'Azur (until December).
- T. Papadopoulo is a also member of the Neuromod scientific council and represents Neuromod in the EUR Healthy (EUR: Universitary Research School) and in the Maison de la Simulation of University Côte d'Azur.
9.2 Teaching - Supervision - Juries
9.2.1 Teaching
- Master: T. Papadopoulo, Inverse problems for brain functional imaging, 3h, M2, Mathématiques, Vision et Apprentissage, ENS Cachan, France.
- Master: T. Papadopoulo and S. Deslauriers-Gauthier Functional Brain Imaging, each 15h, M1,M2 in the MSc Mod4NeuCog of Université Côte d'Azur.
- Master: T. Papadopoulo and S. Deslauriers-Gauthier, Application of machine learning to MRI, electophysiology & brain computer interfaces, each 10h, M1,M2 in the MSc Data Science and Artificial Intelligence of Université Côte d'Azur.
- Master: R. Veltz, Mathematical methods for neuroscience, 24h, M2, Mathématiques, Vision et Apprentissage, ENS Cachan, France.
- Master: S. Deslauriers-Gauthier and Romain Veltz Introduction to Python each 15h, M1 in the MSc Mod4NeuCog of Université Côte d'Azur.
9.2.2 Supervision
- PhD defended on October: I. Carrara, “Auto-regressive models for MEG/EEG processing”, started in Oct. 2021. Supervisor: Théodore Papadopoulo 18.
- PhD in progress: Y. Aeschlimann, ”Brain networks from simultaneous modelling of functional MRI and diffusion MRI”, started in Oct. 2022. Supervisors: Samuel Deslauriers-Gauthier, Théodore Papadopoulo.
- PhD in progress: P. Isan, ”Cerebral eletrophysiological exploitation of in vivo evoked potentials to guide the resection of adult brain tumors”, started in Oct. 2023. Supervisor: F. Almairac. Co-Supervisors: T. Papadopoulo, S. Deslauriers-Gauthier.
- PhD in progress: E. Manka, ”Modeling of evoked potentials by direct current stimulation and their links to electromyography”, started in Oct. 2024. Supervisors: S. Deslauriers-Gauthier, T. Papadopoulo.
- PhD in progress: L. Gee, ”Exploring and exploiting the new capabilities of room temperature MEG sensors”', started in Dec. 2024. Supervisors: T. Papadopoulo, C.Bénar (INSERM, Marseille).
- M2 Emeline Manka, from MSc M4NC in Université Côte d'Azur, ”Modeling of the stimulation artefact during brain direct electrostimulation”, March-August 2024. Supervisor: S. Deslauriers-Gauthier.
- M2 Esin Atkay, from MSc M4NC in Université Côte d'Azur, ”Bifurcation theory of whole brain models applied to Cortical Transitions from Wakefulness to Sleep States”, started in Sep. 2024. Supervisor: R. Veltz.
- M2 Teodora-Lucia Stei, from MSc M4NC in Université Côte d'Azur, ”Predicting task-related functional connectivity from structural connectivity”, started in Sep. 2024. Supervisor: S. Deslauriers-Gauthier.
- M2 Gregorio Rebecchi, from MSc M4NC in Université Côte d'Azur, ”Synaptic inference and plasticity manipulation in neurospheres”', March-August 2024. Supervisors: J.-M. Comby, P. Reynaud-Bouret and R. Veltz.
9.2.3 Juries
- T. Papadopoulo participated in the PhD jury of I. Carrara at Université Côte d'Azur on Oct. 18th, 2024.
- T. Papadopoulo participated in the PhD jury of F. Schlosser-Perrin at Université de Montpellier on Dec. 19th, 2024.
- S. Deslauriers-Gauthier participated in the PhD jury of L. Smolders at Eindhoven University of Technology on Jan. 6th, 2025.
- R. Veltz participated in the PhD jury of Paul Paragot at Université Côte d'Azur on June 11th, 2024.
- R. Veltz defended his HDR 19 entitled “Applications of jump processes to neuroscience” on Oct. 1st, 2025.
- R. Veltz and S. Deslauriers-Gauthier served in the master jury of Mod4NeuCog, Université Côte d'Azur.
9.2.4 Participation in Live events
- R. Veltz. Conference Terra Numerica “Comment se creuser les neurones ?”, January 2024
- R. Veltz. Semaine du cerveau, “Briques élémentaires de mémoire”, March 2024
- R. Veltz. Conference at "Fête de la science", “Briques élémentaires de mémoire”, October 2024
10 Scientific production
10.1 Major publications
- 1 articleStructural connectivity to reconstruct brain activation and effective connectivity between brain regions.Journal of Neural Engineering173June 2020, 035006HALDOI
- 2 articleConsensus Matching Pursuit for multi-trial EEG signals.Journal of Neuroscience Methods1801May 2009, 161-170HALDOIback to text
- 3 articleWhite Matter Information Flow Mapping from Diffusion MRI and EEG.NeuroImageJuly 2019HALDOIback to textback to text
- 4 articleA Unified Framework for Multimodal Structure-function Mapping Based on Eigenmodes.Medical Image AnalysisAugust 2020, 22HALDOIback to text
- 5 articleAdaptive Waveform Learning: A Framework for Modeling Variability in Neurophysiological Signals.IEEE Transactions on Signal Processing65August 2017, 4324 - 4338HALDOIback to text
- 6 articleA common formalism for the Integral formulations of the forward EEG problem.IEEE Transactions on Medical Imaging241January 2005, 12-28HALDOIback to text
- 7 articleA Myoelectric Digital Twin for Fast and Realistic Modelling in Deep Learning.Nature Communications141March 2023, 1600HALDOI
10.2 Publications of the year
International journals
International peer-reviewed conferences
Conferences without proceedings
Doctoral dissertations and habilitation theses
Reports & preprints
Other scientific publications
10.3 Cited publications
- 32 articleClassification of covariance matrices using a Riemannian-based kernel for BCI applications.Neurocomputing112July 2013, 172-178HALDOIback to text
- 33 articleStructural connectivity to reconstruct brain activation and effective connectivity between brain regions.Journal of Neural Engineering173June 2020, 035006HALDOIback to text
- 34 articleRelationship Between Flow and Metabolism in BOLD Signals: Insights from Biophysical Models.Brain Topography2412011, 40--53URL: http://dx.doi.org/10.1007/s10548-010-0166-6DOIback to text
- 35 inproceedingsMultivariate Convolutional Sparse Coding for Electromagnetic Brain Signals.Advances in Neural Information Processing Systems31Curran Associates, Inc.2018, URL: https://proceedings.neurips.cc/paper/2018/file/64f1f27bf1b4ec22924fd0acb550c235-Paper.pdfback to text
- 36 articleAdaptive Waveform Learning: A Framework for Modeling Variability in Neurophysiological Signals.IEEE Transactions on Signal Processing6516April 2017, 4324--4338HALDOIback to text
- 37 articleA Riemannian Network for SPD Matrix Learning.Proceedings of the AAAI Conference on Artificial Intelligence311Feb. 2017, URL: https://ojs.aaai.org/index.php/AAAI/article/view/10866DOIback to text
- 38 articleMOABB: trustworthy algorithm benchmarking for BCIs.Journal of neural engineering1562018, 066011back to text
- 39 inproceedingsConnectivity-informed M/EEG inverse problem.GRAIL 2020 - MICCAI Workshop on GRaphs in biomedicAl Image anaLysisLima, PeruOctober 2020HALback to text
- 40 inproceedingsIncorporating transmission delays supported by diffusion MRI in MEG source reconstruction.ISBI 2021 - IEEE International Symposium on Biomedical ImagingNice, FranceApril 2021HALback to text
- 41 articleAs above, so below? Towards understanding inverse models in BCI.Journal of Neural Engineering151December 2017back to text
- 42 articleSource localization using recursively applied and projected (RAP) MUSIC.IEEE Transactions on Signal Processing472February 1988, 332 -- 340DOIback to text
- 43 articleOptimal state-space reconstruction using derivatives on projected manifold.Physical Review E8722013, 022905back to text
- 44 incollectionDetecting strange attractors in turbulence.Dynamical systems and turbulence, Warwick 1980Springer1981, 366–381back to text

