Section: New Software and Platforms
Platforms
Propag-5
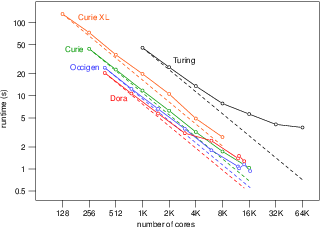
Applied modeling studies performed by the Carmen team, especially M. Potse and M. Kania, in collaboration with IHU Liryc and foreign partners [39], [43], [18], [20], [14] [45], rely to a great extent on high-performance computations on the national supercomputers Curie, Occigen, and Turing. While the newly developed CEPS code is not ready to run efficiently on these systems we rely on an older code named Propag-5. This code is the result of a decades-long development first at the Université de Montréal in Canada, then at Maastricht University in the Netherlands, and finally at the Institute of Computational Science of the Università della Svizzera italiana in Lugano, Switzerland. Relatively small contributions to this code have been made by the Carmen team.
The predecessor of Propag-5, named Propag-4, was developed by M. Potse at the Université de Montréal [8]. It was based on earlier model code developed there by the team of Prof. R. Gulrajani [50], [53], and was parallellized with OpenMP to utilize the shared-memory SGI supercomputers available there at the time. Propag-4 was the first code ever able to run a bidomain reaction-diffusion model of the entire human ventricles; a problem 30 times larger than what had been reported before [8].
In order to utilize the more recent distributed-memory architectures Propag-4 was transformed into the hybrid MPI-OpenMP code Propag-5 at the Institute of Computational Science in Lugano by D. Krause and M. Potse [49]. The resulting code has been used for numerous applied studies. An important limitation of the Propag code is that it relies on a semi-structured mesh with a uniform resolution. On the other hand, the code scales excellently to large core counts and, as it is controlled completely with command-line flags and configuration files, it can be used by non-programmers. It also features
-
a completely parallel workflow, including the initial anatomy input and mesh partitioning, which allows it to work with meshes of more than nodes,
-
a flexible output scheme allowing hundreds of different state variables and transient variables to be output to file, when desired, using any spatial and temporal subsampling,
-
a configurable, LUSTRE-aware parallel output system in which groups of processes write HDF5/netCDF files, and
The code has been stable and reliable for several years, and only minor changes are being made currently. It can be considered the workhorse for our HPC work until CEPS takes over.
Gepetto
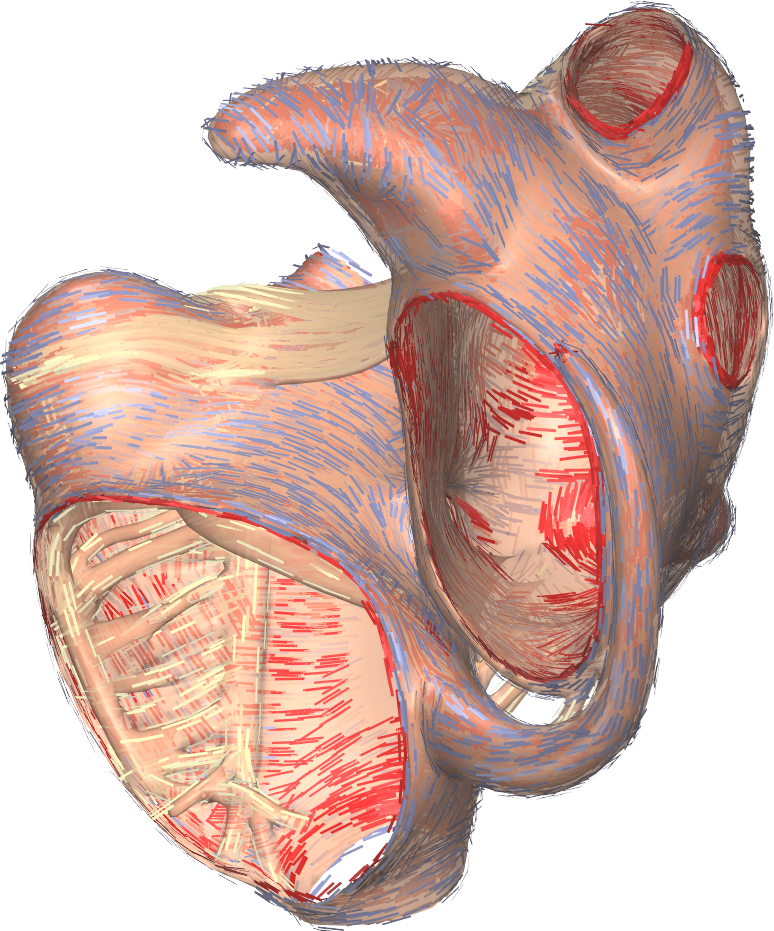
Gepetto, named after a famous model maker, is a sofware suite that transforms a surface mesh of the heart into a set of (semi-)structured meshes for use by the Propag software or others. It creates the different fiber orientations in the model, including the transmurally rotating ventricular fibers and the various bundle structures in the atria (figure 2), and creates layers with possibly different electrophysiological properties across the wall. A practically important function is that it automatically builds the matching heart and torso meshes that Propag uses to simulate potentials in the torso (at a resolution of 1 mm) after projecting simulation results from the heart model (at 0.1 to 0.2 mm) on the coarser torso mesh [52]. Like Propag, the Gepetto software results from a long-term development that started in Montreal, Canada, around 2002. The code for atrial fiber structure was developed by our team.
|
MUSIC
MUSIC is a multimodal platform for cardiac imaging developed by the imaging team at IHU LIRYC in collaboration with the Inria team Asclepios (https://bil.inria.fr/fr/software/view/1885/tab). It is based on the medInria software also developed by the Asclepios team. MUSIC is a cross-platform software for segmentation of medical imaging data, meshing, and ultimately also visualization of functional imaging data and model results.
Several members of the Carmen team use MUSIC for their work. The team also contributes a series of plugins for MUSIC through the IDAM project (section 6.2).