Section: New Results
DimSUM: Dimension and Scale Unifying Maps for Visual Abstraction of DNA Origami Structures
Participants : Haichao Miao [TU Wien, Austria, and Austrian Institute of Technology, Vienna, Austria] , Elisa de Llano [Austrian Institute of Technology, Vienna, Austria] , Tobias Isenberg [correspondant] , M. Eduard Gröller [TU Wien, Austria] , Ivan Barišic [Austrian Institute of Technology, Vienna, Austria] , Ivan Viola [TU Wien, Austria and KAUST, Kingdom of Saudi Arabia] .
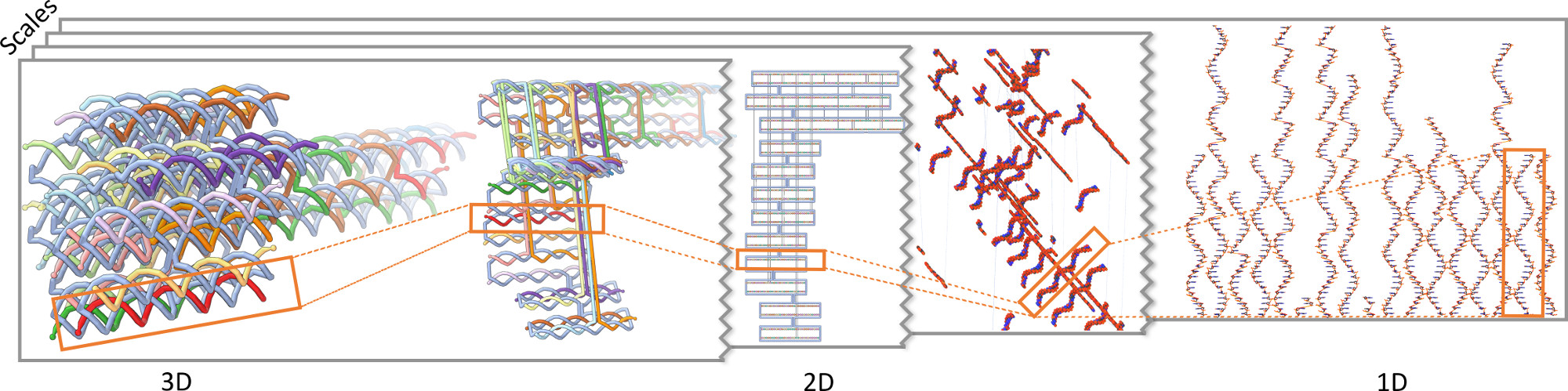
We presented a novel visualization concept for DNA origami structures that integrates a multitude of representations into a DimSUM. This novel abstraction map (see Figure 9) provides means to analyze, smoothly transition between, and interact with many visual representations of the DNA origami structures in an effective way that was not possible before. DNA origami structures are nanoscale objects, which are challenging to model in silico. In our holistic approach we seamlessly combined three-dimensional realistic shape models, two-dimensional diagrammatic representations, and ordered alignments in one-dimensional arrangements, with semantic transitions across many scales. To navigate through this large, two-dimensional abstraction map we highlighted locations that users frequently visit for certain tasks and datasets. Particularly interesting viewpoints can be explicitly saved to optimize the workflow. We have developed DimSUM together with domain scientists specialized in DNA nanotechnology. In the paper we discussed our design decisions for both the visualization and the interaction techniques. We demonstrateed two practical use cases in which our approach increases the specialists' understanding and improves their effectiveness in the analysis. Finally, we discussed the implications of our concept for the use of controlled abstraction in visualization in general.
More on the project Web page: https://tobias.isenberg.cc/VideosAndDemos/Miao2018DDS.