Section: New Results
Imaging
ML estimation of wavelet regularization hyperparameters in inverse problems
Participant : Laure Blanc-Féraud.
This work was made in collaboration with Caroline Chaux from LATP (Marseille) and Roberto Cavicchioli and Luca Zanni from University of Modena (Italy).
Parameter estimation, Maximum likelihood estimation, Wavelet transforms, Deconvolution, Gradi- ent methods
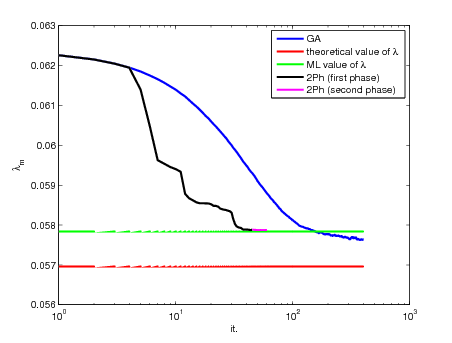
We are interested in regularizing hyperparameter estimation by maximum likelihood in inverse problems with wavelet regularization. One parameter per subband is estimated by gradient ascent algorithm.We have to face with two main difficulties: i) sampling the a posteriori image distribution to compute the gradient of the objective function; ii) choosing a suited step-size to ensure good convergence properties of the gradient ascent algorithm. We first show that introducing an auxiliary variable makes the sampling feasible using classical Metropolis-Hastings algorithm and Gibbs sampler. Secondly, we propose an adaptive step-size selection and a line-search strategy to improve the gradient-based method. Good performances of the proposed approach are demonstrated on both synthetic and real data.
|
Joint optimization of noisy image coding and denoising
Participants : Mikael Carlavan, Laure Blanc-Féraud.
this work was made in collaboration with Marc Antonini (I3S), Roberto Camarero and Christophe Latry (CNES) and Yves Bobichon (TAS).
coding, denoising, wavelet transform, global rate-distortion optimization
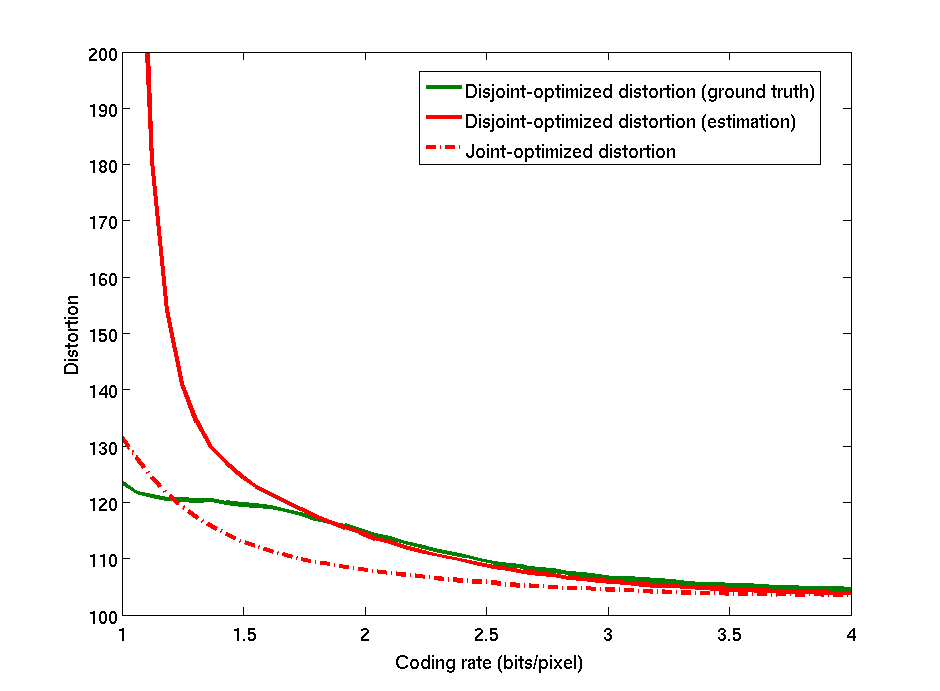
This work concerns the study of optimal noisy source coding/denoising. A global optimization of the problem is usually difficult to perform as the global fidelity criterion needs to be optimized in the same time over the sets of both coding and denoising parameters. Most of the bibliography in this domain is based on the fact that, for a specific criterion, the global optimization problem can be simply separated into two independent optimization problems: The noisy image should be first optimally denoised and this denoised image should then be optimally coded. In many applications however, the layout of the acquisition imaging chain is fixed and can not be changed, that is a denoising step can not be inserted before coding. For this reason, we are concerned here with the problem of global joint optimization in the case the denoising step is performed, as usual, after coding/decoding. In this configuration, we showed on a simple case how to express the global distortion as a function of the coding and denoising parameters. We presented an algorithm to minimize this distortion to get the optimal values of these parameters. Figure 2 shows results of this joint optimization algorithm, on the classical test image Barbara, in comparison to the usual disjoint optimization technique, which consists in selecting the coding and the denoising parameters such that the coding and the denoising errors are independently minimized. On the range of validity of the proposed model, we see that the joint optimized distortion slightly outperforms the disjoint optimized distortion (in the presented example, the PSNR of the reconstructed image increases of at bits/pixels). The interesting point of the proposed method is that it allows to reach the same global error than the disjoint optimized technique but for a lower coding rate. For example, on this image, the joint optimization technique reaches at bits/pixel the same distortion than the one obtained at bits/pixels for the disjoint optimization technique. The benefit in terms of compression performances of the joint optimization appears then to be very significant.
|
Blind deconvolution
Participants : Saima Ben Hadj, Laure Blanc-Féraud.
This research takes place within the ANR DIAMOND. This work was made in collaboration with Gilles Aubert, Laboratoire J. Dieudonné (CNRS,UNS).
One of our tasks within the ANR Diamond project is the blind restoration of images coming from Confocal laser scanning microscopy (CLSM). CLSM is a powerful technique for studying biological specimens in three dimensions by optical sectioning. Nevertheless, it suffers from some artifacts. First, CLSM images are affected by a depth-variant (DV) blur due to spherical aberrations induced by refractive index mismatch between the different media composing the system as well as the specimen. Second, CLSM images are corrupted with a Poisson noise due to low illumination. Because of these intrinsic optical limitations, it is essential to remove both DV blur and noise from these images by digital processing.
In this context, we first study space-variant (SV) blur models and prove that a model where the SV point spread function (PSF) is approximated by a convex combination of a set of space-invariant (SI) PSFs is efficient and adequate to the inversion problem [30] [10] . Afterwards, we focus on the non-bind restoration problem and we fit a fast restoration method based on a domain decomposition technique [33] to our DV blur model [10] , [9] .
Recently, we focus on the blind case. In fact, in practice it is difficult to obtain the DV PSF in spite of the existence of theoretical PSF models [34] , because these models are dependent on some unknown acquisition parameters (e.g. the refractive index (RI) of the specimen). Therefore a blind or semi-blind restoration algorithm is needed for this system. We propose two methods for this problem : In the first method, we define a criterion to be jointly minimized w.r.t to the image and the PSF set. In this method, the intensities of each SI PSF are estimated at every voxel. Although the big number of parameters to be estimated, the method allows more freedom on the shape of the PSF which could be more or less deformed according to spherical aberration level. We provide a theoretical proof of the existence of a minimizer of the considered problem [23] . Then, we perform the minimization by following an alternate minimization scheme, each elementary minimization is performed using the recently proposed scaled gradient projection (SGP) algorithm that has shown a fast convergence rate [29] . Results on simulated CLSM images and comparison with another alternate scheme based on a regularized version of the Richardson–Lucy algorithm [31] are shown in Fig. 3 . In the second blind method, we use a Gaussian approximation of each of the SI PSFs. This presents the advantage of significantly reducing the number of parameters to be estimated but constraints the PSF shape. We prove on simulated data that the first method provides more accurate restoration result than the second one.
|
Morphogenesis of living organisms
Participant : Grégoire Malandain.
This research takes place within the Inria Large-scale initiative Morphogenetics.
This work was made in collaboration with Christophe Godin and Léo Guignard from Virtual Plants.
super-resolution, SPIM, morphogenesis
We extended a previous work [32] for the reconstruction of microscopic images. In particular, we extended the super-resolution image reconstruction (where several images, acquired from different viewpoints, are fused) to the lightsheet (or SPIM for Selective Plane Illumination Microscope) microscope modality. This modality offers a high acquisition speed, allowing imaging an organism frequently. As an exemple, Phallusia mammillata and Ciona intestinalis embryos can be imaged from 32 cells to around 1000 cells. The organism is captured from four different angles every 2 minutes during 2 hours (collaboration with CRBM Montpellier and EMBL Heidelberg).